"clathrin, light chain A"
ZFIN 
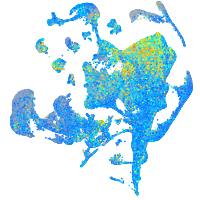
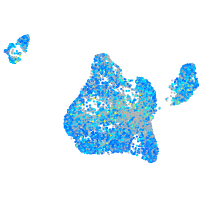
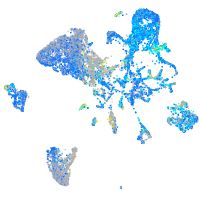
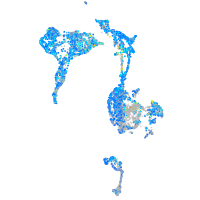
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| actb1 | 0.456 | bhmt | -0.372 |
| cldn15la | 0.437 | aqp12 | -0.335 |
| anxa2b | 0.422 | agxtb | -0.334 |
| serpinb1l3 | 0.421 | fetub | -0.297 |
| calm2b | 0.420 | apoa2 | -0.292 |
| cfl1 | 0.414 | rbp4 | -0.289 |
| GCA | 0.414 | apoc1 | -0.286 |
| calm2a | 0.407 | ces2 | -0.282 |
| eps8l3b | 0.404 | fabp10a | -0.281 |
| calml4a | 0.401 | pnp4b | -0.279 |
| vil1 | 0.401 | gatm | -0.276 |
| si:dkeyp-73b11.8 | 0.401 | LOC110437731 | -0.276 |
| wu:fb59d01 | 0.401 | ttr | -0.276 |
| sri | 0.394 | serpina1 | -0.273 |
| myl12.1 | 0.390 | zgc:123103 | -0.272 |
| calm3b | 0.389 | gc | -0.271 |
| arpc3 | 0.389 | comtd1 | -0.270 |
| anks4b | 0.386 | gamt | -0.270 |
| tmem176l.2 | 0.382 | apom | -0.268 |
| si:ch211-137i24.10 | 0.381 | rbp2b | -0.266 |
| zgc:172079 | 0.381 | tfa | -0.265 |
| cotl1 | 0.378 | kng1 | -0.265 |
| cst14a.2 | 0.376 | grhprb | -0.261 |
| ywhaqa | 0.376 | serpina1l | -0.260 |
| cd63 | 0.375 | fabp3 | -0.258 |
| zgc:77748 | 0.374 | ambp | -0.258 |
| s100a10a | 0.372 | eif4ebp3 | -0.257 |
| bin2a | 0.366 | si:dkey-86l18.10 | -0.255 |
| slc9a3r1a | 0.365 | mat1a | -0.255 |
| tmem45b | 0.365 | f7i | -0.254 |
| gnpda1 | 0.362 | fgg | -0.253 |
| serpinb1 | 0.361 | c9 | -0.252 |
| atp5if1a | 0.360 | fgb | -0.250 |
| nccrp1 | 0.359 | hao1 | -0.248 |
| ush1c | 0.359 | prss1 | -0.248 |