chloride intracellular channel 1
ZFIN 
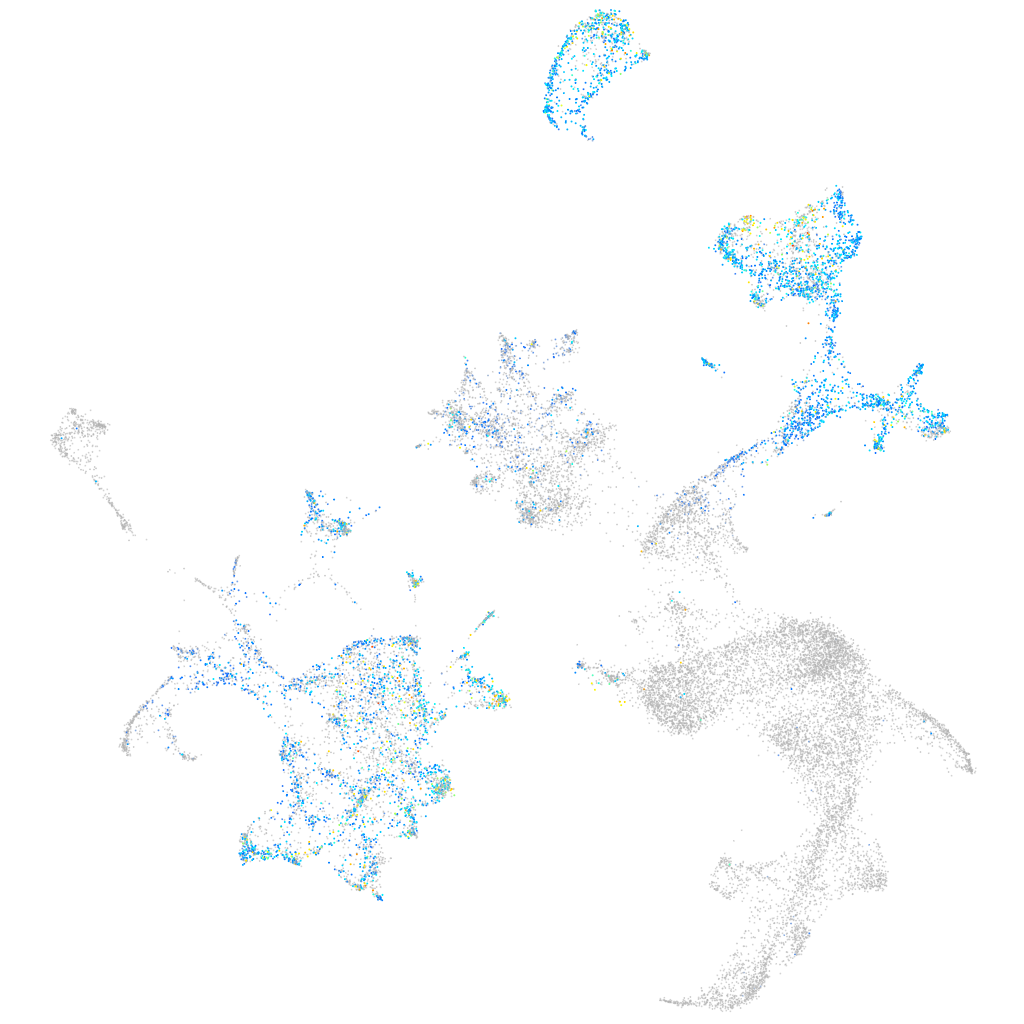
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cotl1 | 0.482 | hbae1.1 | -0.447 |
| cd63 | 0.467 | hbae3 | -0.442 |
| tmsb4x | 0.465 | hbae1.3 | -0.442 |
| actb2 | 0.463 | hbbe1.3 | -0.440 |
| coro1a | 0.458 | hbbe2 | -0.434 |
| arpc1b | 0.456 | hbbe1.1 | -0.419 |
| rac2 | 0.452 | cahz | -0.408 |
| arhgdig | 0.450 | hbbe1.2 | -0.381 |
| arpc3 | 0.447 | hemgn | -0.380 |
| arpc2 | 0.447 | fth1a | -0.378 |
| pfn1 | 0.444 | blvrb | -0.366 |
| cebpb | 0.440 | alas2 | -0.361 |
| actb1 | 0.436 | si:ch211-250g4.3 | -0.340 |
| eno3 | 0.431 | slc4a1a | -0.339 |
| srgn | 0.429 | creg1 | -0.335 |
| lasp1 | 0.422 | nt5c2l1 | -0.331 |
| laptm5 | 0.420 | zgc:163057 | -0.320 |
| cap1 | 0.416 | epb41b | -0.316 |
| cfl1 | 0.412 | tspo | -0.307 |
| zgc:162730 | 0.408 | si:ch211-207c6.2 | -0.300 |
| capgb | 0.408 | nmt1b | -0.290 |
| arpc4l | 0.407 | zgc:56095 | -0.288 |
| gapdhs | 0.405 | prdx2 | -0.282 |
| rhogb | 0.395 | tmod4 | -0.277 |
| sat1a.2 | 0.394 | plac8l1 | -0.264 |
| anxa11b | 0.392 | hbae5 | -0.257 |
| arpc4 | 0.392 | sptb | -0.249 |
| myl12.1 | 0.392 | tfr1a | -0.248 |
| abracl | 0.391 | hbbe3 | -0.244 |
| xbp1 | 0.389 | si:ch211-227m13.1 | -0.243 |
| crip1 | 0.389 | uros | -0.240 |
| nme2b.1 | 0.386 | rfesd | -0.238 |
| GCA | 0.386 | znfl2a | -0.231 |
| capza1b | 0.385 | klf1 | -0.229 |
| pycard | 0.384 | add2 | -0.227 |