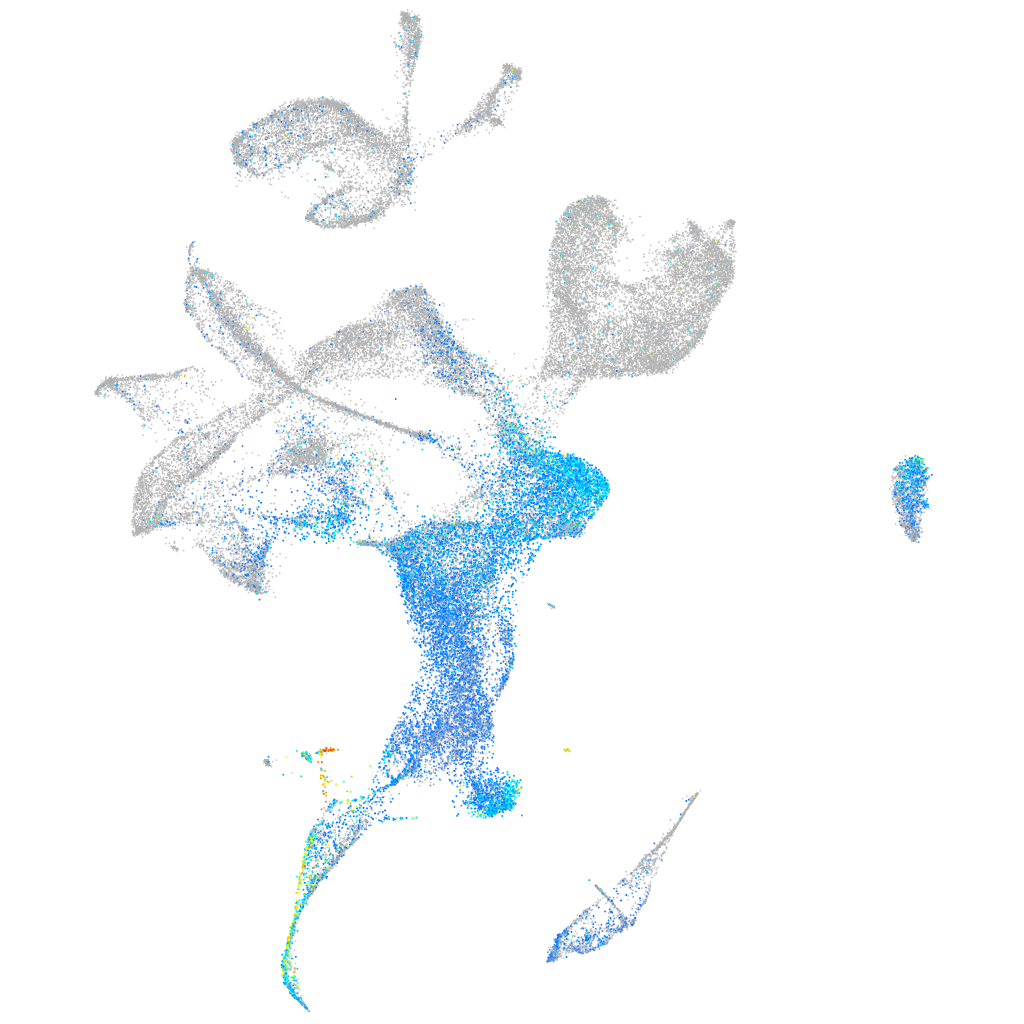
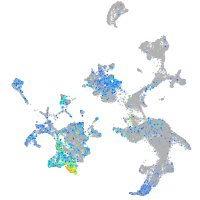
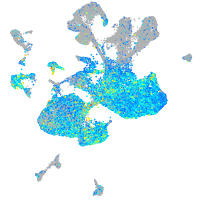
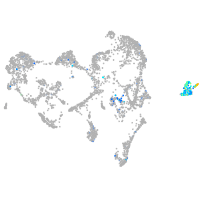
claudin 5a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msna | 0.439 | ckbb | -0.373 |
| tuba8l4 | 0.399 | snap25b | -0.329 |
| lbr | 0.382 | atp6v0cb | -0.327 |
| tuba8l | 0.377 | gapdhs | -0.311 |
| rrm1 | 0.372 | syt5b | -0.298 |
| zgc:56493 | 0.349 | rnasekb | -0.292 |
| mdka | 0.345 | sypb | -0.285 |
| ccna2 | 0.344 | ndrg4 | -0.267 |
| mki67 | 0.343 | atpv0e2 | -0.265 |
| pcna | 0.339 | crx | -0.264 |
| vamp3 | 0.335 | vamp2 | -0.261 |
| tspan7 | 0.332 | rs1a | -0.253 |
| banf1 | 0.330 | atp6v1e1b | -0.253 |
| her15.1 | 0.329 | atp1a3a | -0.241 |
| tuba1b | 0.328 | atp1b2a | -0.240 |
| mcm7 | 0.323 | ppp1r14c | -0.237 |
| ahcy | 0.322 | atp6v1g1 | -0.234 |
| ranbp1 | 0.316 | gng13b | -0.233 |
| anp32b | 0.313 | sncb | -0.233 |
| foxn4 | 0.313 | mpp6b | -0.231 |
| aspm | 0.309 | pcbp3 | -0.230 |
| hes2.2 | 0.308 | gng3 | -0.228 |
| selenoh | 0.308 | scrt2 | -0.225 |
| stmn1a | 0.307 | tpi1b | -0.222 |
| dek | 0.303 | calb2b | -0.218 |
| cks1b | 0.303 | gpm6ab | -0.217 |
| plk1 | 0.302 | zc4h2 | -0.216 |
| hmgb2b | 0.301 | otx5 | -0.207 |
| nutf2l | 0.301 | gnb3a | -0.203 |
| hes6 | 0.299 | rtn1a | -0.202 |
| hmgb2a | 0.299 | elmo1 | -0.200 |
| cdc20 | 0.297 | sh3gl2a | -0.200 |
| dlgap5 | 0.297 | pcp4l1 | -0.200 |
| id1 | 0.297 | mid1ip1b | -0.199 |
| COX7A2 (1 of many) | 0.296 | samsn1a | -0.199 |