cellular inhibitor of PP2A
ZFIN 
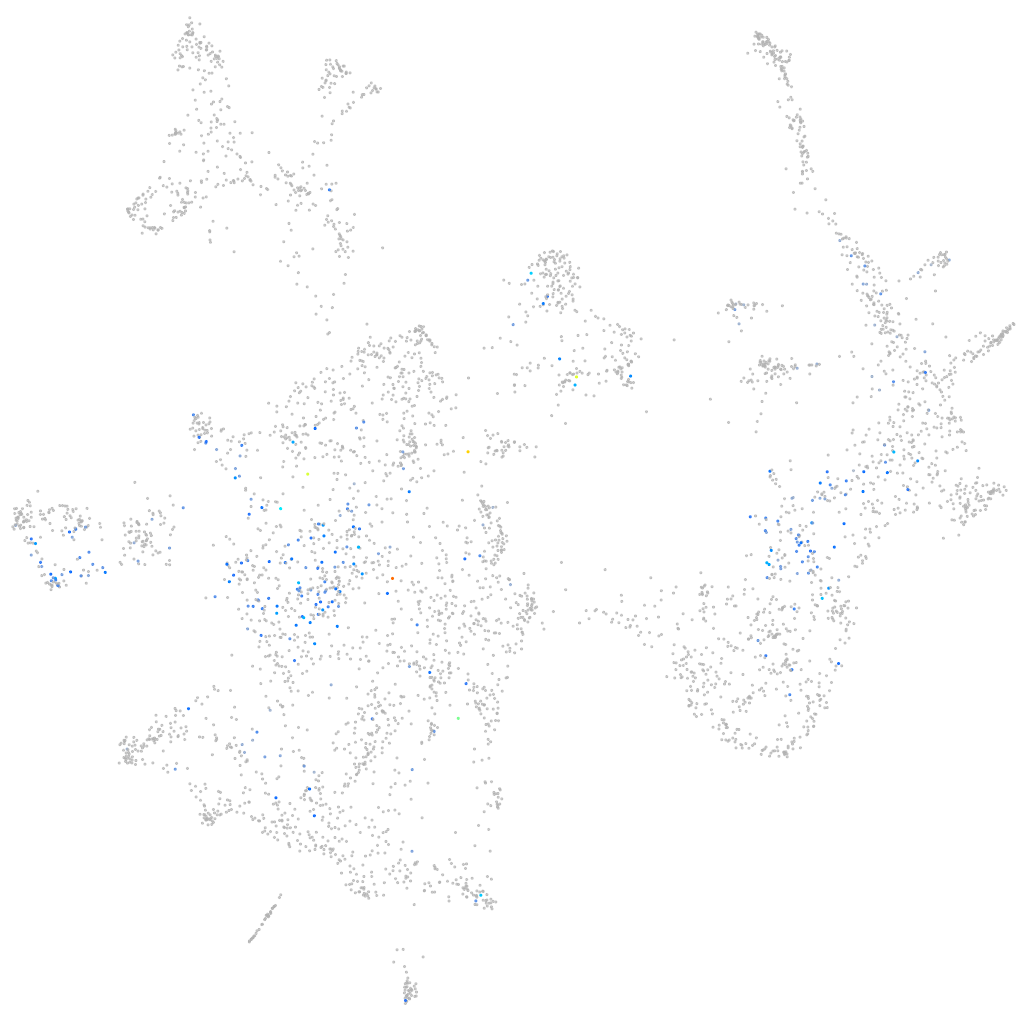
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.330 | gapdhs | -0.090 |
| cdk1 | 0.307 | rnasekb | -0.087 |
| si:dkey-261m9.17 | 0.287 | calm1a | -0.087 |
| ccnb1 | 0.285 | calm1b | -0.084 |
| ncapg | 0.277 | tob1b | -0.082 |
| cks1b | 0.277 | b2ml | -0.082 |
| rrm1 | 0.274 | eno1a | -0.081 |
| ccna2 | 0.269 | atp2b1a | -0.080 |
| mad2l1 | 0.269 | otofb | -0.078 |
| numa1 | 0.266 | gpx2 | -0.078 |
| zgc:165555.3 | 0.265 | atp1a3b | -0.077 |
| tpx2 | 0.264 | osbpl1a | -0.075 |
| zgc:110425 | 0.259 | eef1da | -0.074 |
| aurkb | 0.258 | cd164l2 | -0.074 |
| dlgap5 | 0.257 | calml4a | -0.074 |
| plk1 | 0.256 | pvalb1 | -0.074 |
| zgc:110216 | 0.255 | nptna | -0.074 |
| ncapd2 | 0.253 | CR925719.1 | -0.073 |
| top2a | 0.251 | dedd1 | -0.073 |
| cdca5 | 0.251 | gabarapb | -0.073 |
| zgc:86764 | 0.249 | ucp2 | -0.072 |
| kif23 | 0.248 | tmem59 | -0.072 |
| armc1l | 0.247 | COX3 | -0.071 |
| cdca8 | 0.246 | itm2ba | -0.071 |
| chaf1a | 0.246 | nupr1a | -0.071 |
| nusap1 | 0.246 | map1lc3a | -0.071 |
| prc1a | 0.246 | si:dkey-16p21.8 | -0.070 |
| hmgb2a | 0.243 | si:ch73-15n15.3 | -0.070 |
| si:ch211-113a14.24 | 0.242 | emb | -0.070 |
| birc5a | 0.242 | pvalb8 | -0.070 |
| mibp | 0.242 | id2a | -0.069 |
| stmn1a | 0.242 | gabarapa | -0.069 |
| ube2c | 0.241 | gb:eh456644 | -0.069 |
| pbk | 0.241 | tpi1a | -0.069 |
| fbxo5 | 0.240 | atp1b2b | -0.068 |