cytosolic iron-sulfur assembly component 1
ZFIN 
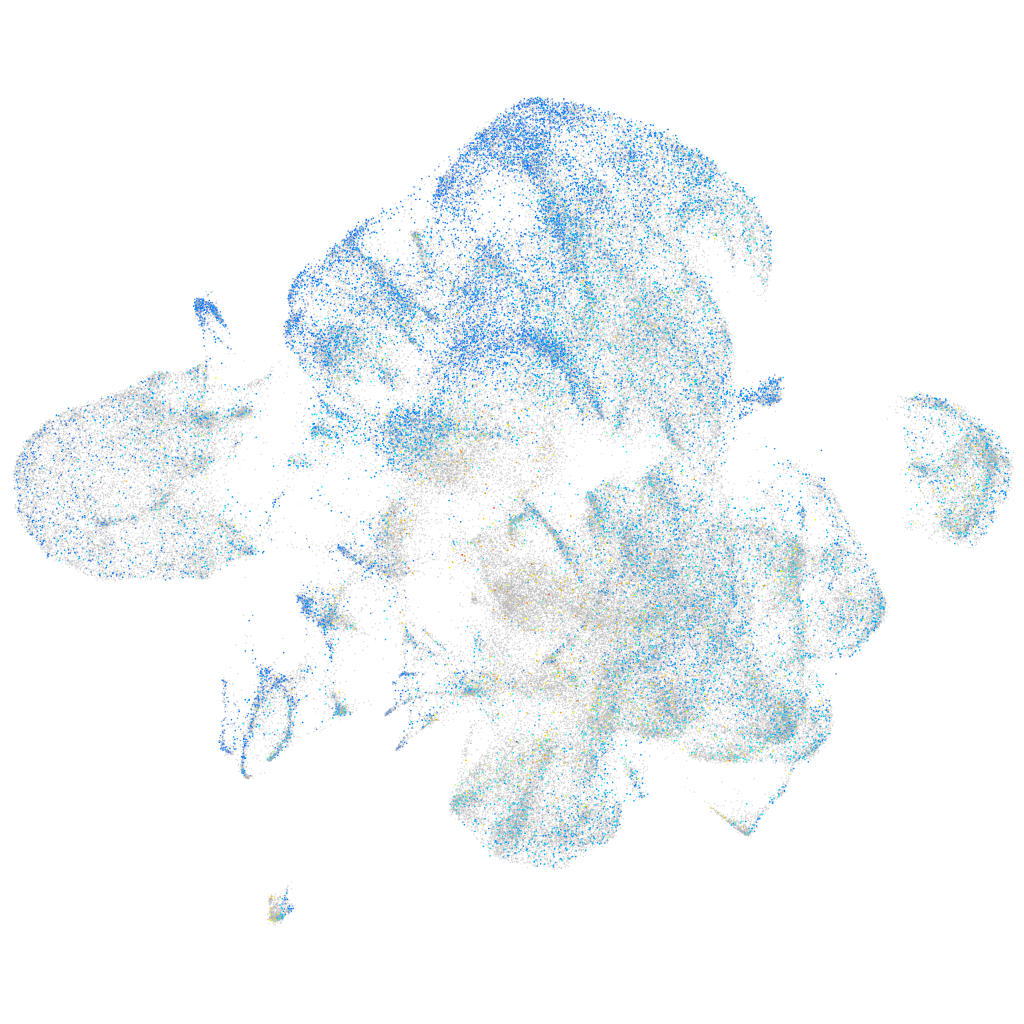
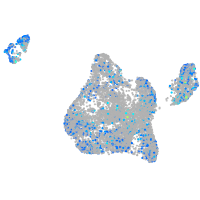
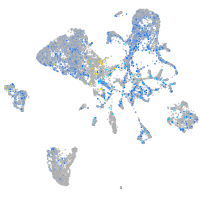
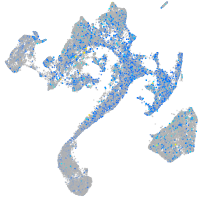
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.117 | NC-002333.4 | -0.051 |
| zgc:56493 | 0.110 | gpm6ab | -0.046 |
| rrm2 | 0.105 | gpm6aa | -0.044 |
| rrm1 | 0.103 | rtn1a | -0.044 |
| pcna | 0.099 | stm | -0.040 |
| rpa2 | 0.098 | COX3 | -0.039 |
| ran | 0.098 | si:ch211-152c2.3 | -0.038 |
| tuba8l4 | 0.096 | elavl3 | -0.037 |
| cks1b | 0.095 | pvalb2 | -0.036 |
| eif3ba | 0.095 | apoc1 | -0.036 |
| h3f3a | 0.095 | pou5f3 | -0.035 |
| nutf2l | 0.094 | stmn1b | -0.035 |
| pa2g4b | 0.093 | elavl4 | -0.035 |
| tuba8l | 0.091 | pvalb1 | -0.034 |
| rnaseh2a | 0.091 | rtn1b | -0.034 |
| slc25a3b | 0.090 | atp6v0cb | -0.034 |
| cnbpa | 0.090 | crabp2b | -0.034 |
| rplp0 | 0.090 | COX7A2 | -0.034 |
| ranbp1 | 0.089 | aplp1 | -0.033 |
| mcm7 | 0.089 | adgrl3.1 | -0.033 |
| rpl7 | 0.089 | serinc1 | -0.031 |
| ccna2 | 0.088 | nsg2 | -0.031 |
| hnrnpa0l | 0.088 | atp1a3a | -0.031 |
| shmt1 | 0.087 | mt-co2 | -0.031 |
| rfc5 | 0.087 | myt1la | -0.031 |
| selenoh | 0.086 | sncb | -0.030 |
| ptgr1 | 0.086 | stx1b | -0.030 |
| rpl19 | 0.086 | CR383676.1 | -0.030 |
| cdca5 | 0.086 | actc1b | -0.030 |
| rps7 | 0.085 | sp5l | -0.029 |
| cbx3a | 0.085 | sox4a | -0.029 |
| snrpd1 | 0.084 | LOC108190024 | -0.029 |
| rfc3 | 0.084 | robo2 | -0.029 |
| rpa3 | 0.084 | syt1a | -0.029 |
| rpl8 | 0.084 | rbfox1 | -0.029 |