charged multivesicular body protein 5b
ZFIN 
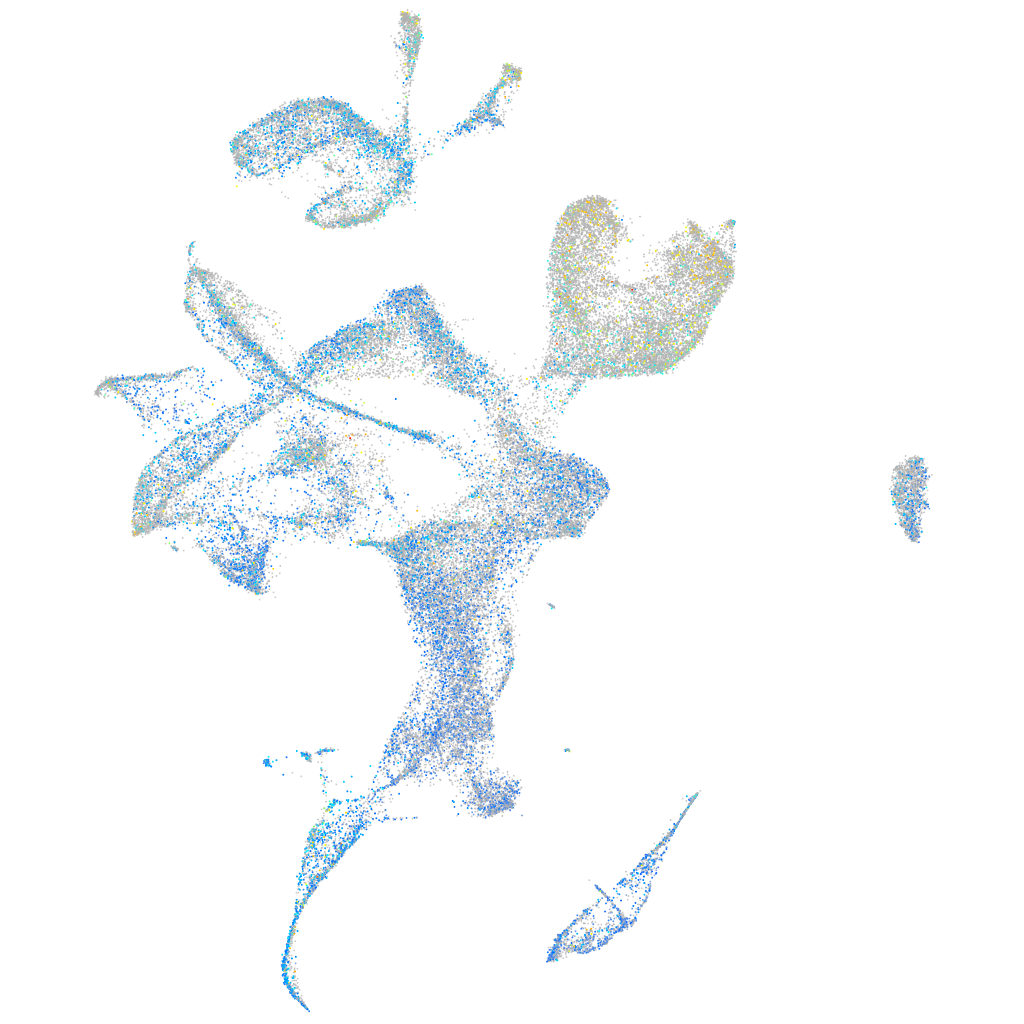
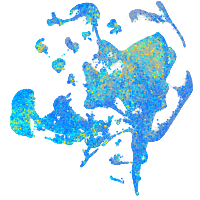
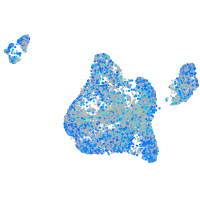
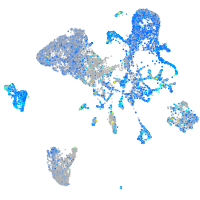
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tspan36 | 0.127 | si:ch73-1a9.3 | -0.072 |
| dct | 0.125 | ptmab | -0.058 |
| slc45a2 | 0.124 | vsx1 | -0.047 |
| gpr143 | 0.124 | neurod4 | -0.044 |
| tyrp1b | 0.123 | hes2.2 | -0.044 |
| gstp1 | 0.122 | hmgb2a | -0.039 |
| pmela | 0.121 | hist1h4l | -0.034 |
| rab38 | 0.118 | hes2.1 | -0.034 |
| sparc | 0.117 | calb2b | -0.034 |
| bace2 | 0.116 | nrn1lb | -0.032 |
| tspan10 | 0.115 | rrm2 | -0.032 |
| tyrp1a | 0.114 | si:ch73-281n10.2 | -0.032 |
| pmelb | 0.111 | CABZ01005379.1 | -0.032 |
| pttg1ipb | 0.110 | zgc:165555.3 | -0.031 |
| qdpra | 0.110 | ccne2 | -0.031 |
| triobpa | 0.109 | si:dkey-261m9.17 | -0.031 |
| cd63 | 0.107 | lig1 | -0.031 |
| oca2 | 0.106 | mdkb | -0.031 |
| rnaseka | 0.106 | LOC798783 | -0.030 |
| pcbd1 | 0.105 | gnb3a | -0.030 |
| fabp11b | 0.104 | krcp | -0.030 |
| cracr2ab | 0.103 | zgc:110425 | -0.030 |
| tmem98 | 0.103 | si:ch211-113a14.18 | -0.030 |
| rab32a | 0.102 | fezf2 | -0.029 |
| rgrb | 0.102 | hmgn2 | -0.029 |
| agtrap | 0.102 | nova2 | -0.029 |
| zgc:110591 | 0.101 | dla | -0.029 |
| comtb | 0.100 | fbxo5 | -0.029 |
| col4a5 | 0.099 | vsx2 | -0.029 |
| rlbp1b | 0.099 | chaf1a | -0.029 |
| slc24a5 | 0.098 | slbp | -0.028 |
| col4a6 | 0.098 | zgc:110216 | -0.028 |
| mitfa | 0.098 | cadm3 | -0.028 |
| fstl1b | 0.097 | esco2 | -0.028 |
| msnb | 0.097 | foxn4 | -0.028 |