chico
ZFIN 
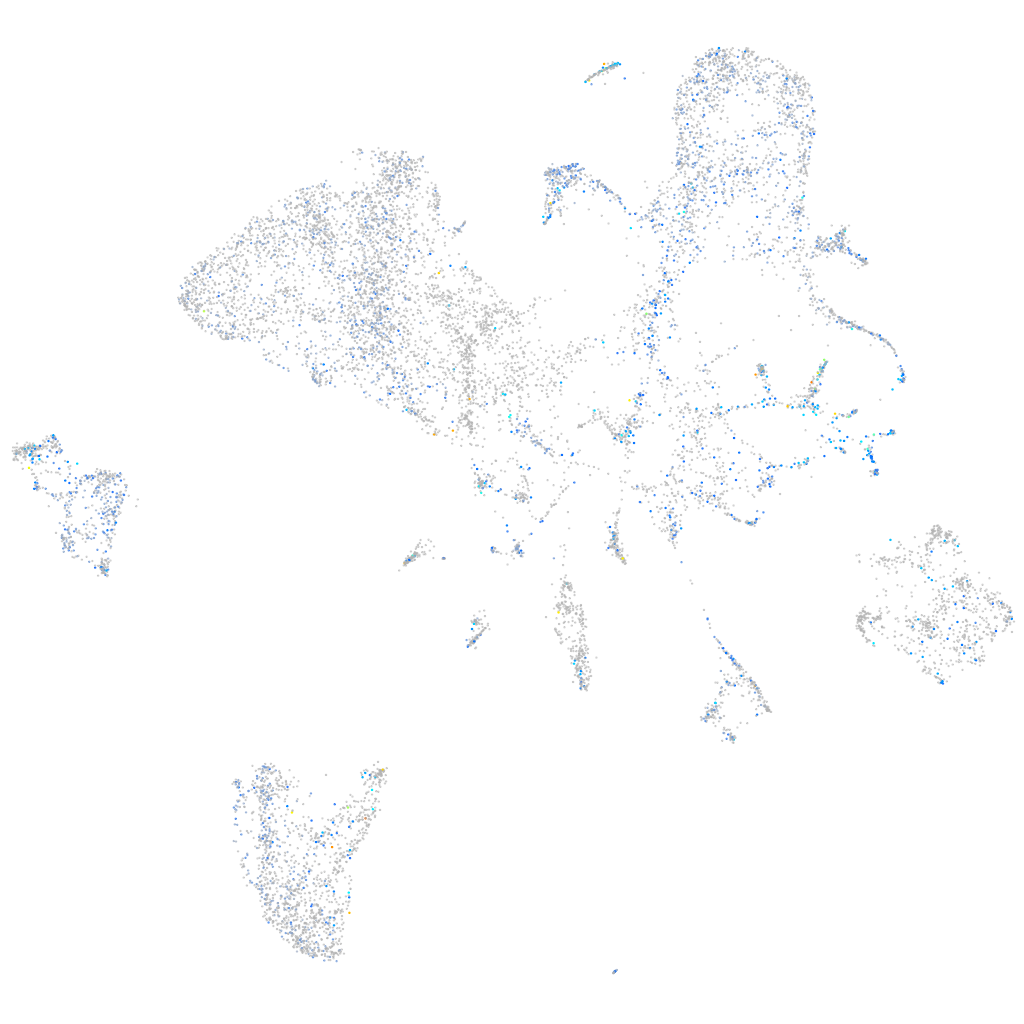
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.199 | apoc1 | -0.133 |
| mir7a-1 | 0.190 | gamt | -0.132 |
| neurod1 | 0.187 | bhmt | -0.127 |
| vat1 | 0.184 | apoa1b | -0.120 |
| pcsk2 | 0.180 | apoa2 | -0.116 |
| si:ch1073-429i10.3.1 | 0.175 | agxtb | -0.116 |
| insm1a | 0.172 | gatm | -0.116 |
| id4 | 0.170 | fabp10a | -0.114 |
| insm1b | 0.170 | zgc:123103 | -0.113 |
| CR383676.1 | 0.170 | gapdh | -0.112 |
| scg2a | 0.167 | fetub | -0.112 |
| gapdhs | 0.166 | serpina1 | -0.111 |
| pcdh20 | 0.164 | rbp2b | -0.111 |
| calm1b | 0.162 | serpina1l | -0.110 |
| scgn | 0.161 | hao1 | -0.110 |
| pcsk1 | 0.161 | apom | -0.109 |
| fev | 0.158 | tfa | -0.109 |
| pcsk1nl | 0.158 | ttr | -0.108 |
| syt1a | 0.155 | ahcy | -0.107 |
| zgc:101731 | 0.155 | apoa4b.1 | -0.107 |
| CABZ01072885.1 | 0.155 | pnp4b | -0.107 |
| calm1a | 0.154 | hpda | -0.106 |
| kdm6bb | 0.152 | LOC110437731 | -0.106 |
| si:dkey-153k10.9 | 0.151 | afp4 | -0.105 |
| slc35g2a | 0.149 | fgg | -0.105 |
| nkx2.2a | 0.147 | rbp4 | -0.105 |
| tmem63c | 0.147 | ces2 | -0.104 |
| gnb1a | 0.146 | ambp | -0.104 |
| vamp2 | 0.144 | uox | -0.103 |
| ier2b | 0.143 | grhprb | -0.103 |
| mir375-2 | 0.142 | agxta | -0.102 |
| nalcn | 0.142 | gc | -0.102 |
| appa | 0.142 | aqp12 | -0.101 |
| rnasekb | 0.142 | si:dkey-86l18.10 | -0.101 |
| capns1b | 0.141 | fgb | -0.100 |