cilia and flagella associated protein 126
ZFIN 
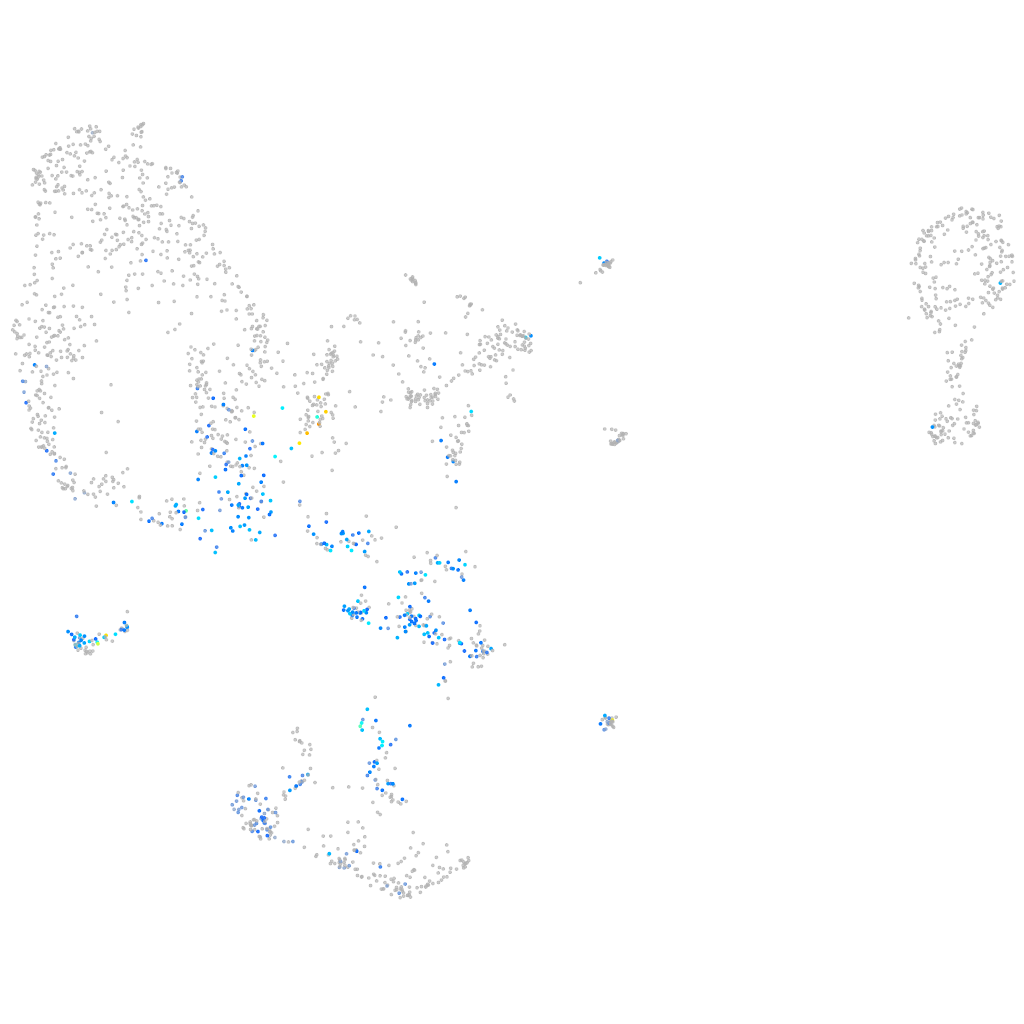
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.481 | gapdh | -0.229 |
| ccdc169 | 0.479 | gcshb | -0.213 |
| capslb | 0.454 | alpl | -0.209 |
| ccdc173 | 0.453 | agxtb | -0.208 |
| tex9 | 0.443 | glud1b | -0.207 |
| si:ch211-163l21.7 | 0.441 | gstt1a | -0.206 |
| foxj1a | 0.440 | epdl2 | -0.206 |
| cfap20 | 0.436 | slc16a1b | -0.203 |
| ribc2 | 0.433 | si:dkey-33i11.9 | -0.203 |
| dnali1 | 0.433 | cx32.3 | -0.203 |
| ccdc151 | 0.433 | phyhd1 | -0.202 |
| spata4 | 0.431 | cox6a2 | -0.200 |
| enkur | 0.429 | dpys | -0.198 |
| si:dkey-70b23.2 | 0.428 | rplp2 | -0.197 |
| spag6 | 0.426 | nipsnap3a | -0.197 |
| tctex1d1 | 0.425 | si:ch211-139a5.9 | -0.194 |
| tekt1 | 0.425 | gpt2l | -0.193 |
| tekt4 | 0.424 | haao | -0.192 |
| efcab1 | 0.421 | g6pca.2 | -0.187 |
| dnal1 | 0.420 | rgn | -0.186 |
| ttc25 | 0.418 | cubn | -0.182 |
| ccdc114 | 0.418 | slc5a12 | -0.181 |
| capsla | 0.418 | lgmn | -0.181 |
| iqcg | 0.412 | cat | -0.181 |
| spaca9 | 0.412 | gstp1 | -0.180 |
| si:ch211-248e11.2 | 0.409 | slc26a6l | -0.180 |
| cd151l | 0.409 | ASS1 | -0.180 |
| mycbp | 0.406 | ctsz | -0.178 |
| LOC101883320 | 0.405 | sod1 | -0.178 |
| pacrg | 0.405 | pnp6 | -0.177 |
| ncs1a | 0.403 | oplah | -0.177 |
| RSPH1 | 0.402 | si:ch211-201h21.5 | -0.177 |
| efcab2 | 0.401 | rpl3 | -0.175 |
| cfap45 | 0.401 | hao1 | -0.174 |
| dynlt1 | 0.400 | aco1 | -0.174 |