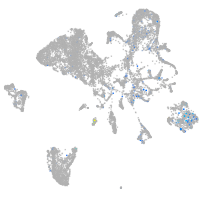
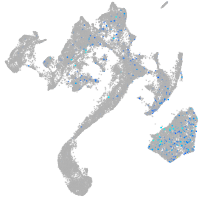
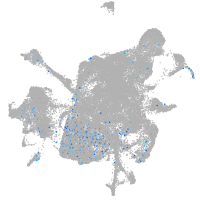
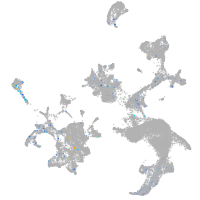
centrosomal protein 152
ZFIN 
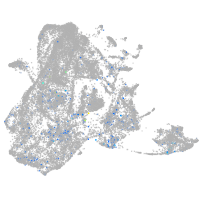
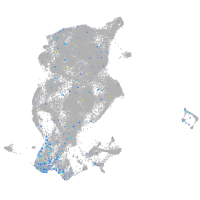
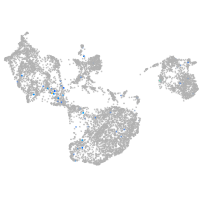
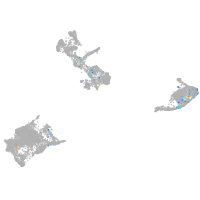
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.205 | paics | -0.099 |
| slc39a10 | 0.202 | CABZ01021592.1 | -0.086 |
| oca2 | 0.191 | impdh1b | -0.081 |
| si:ch73-106l15.4 | 0.188 | uraha | -0.077 |
| slc22a2 | 0.187 | si:dkey-251i10.2 | -0.070 |
| si:ch73-196l6.5 | 0.186 | slc2a15a | -0.068 |
| tyrp1a | 0.184 | glulb | -0.067 |
| kcnj13 | 0.184 | phyhd1 | -0.067 |
| rap1gap | 0.178 | tmem130 | -0.066 |
| prkar1b | 0.177 | atic | -0.063 |
| LOC108190593 | 0.176 | aldob | -0.061 |
| aadac | 0.175 | prps1a | -0.056 |
| mtbl | 0.173 | aox5 | -0.055 |
| pmela | 0.172 | si:ch211-251b21.1 | -0.054 |
| sypl1 | 0.171 | pax7b | -0.053 |
| LOC101885086 | 0.170 | bco1 | -0.052 |
| LOC101882312 | 0.169 | ppat | -0.052 |
| tyrp1b | 0.168 | si:ch211-194k22.8 | -0.052 |
| slc30a1b | 0.166 | cax1 | -0.050 |
| slc6a15 | 0.165 | rpl7 | -0.050 |
| BX088596.2 | 0.163 | rps5 | -0.049 |
| dglucy | 0.162 | akr1b1 | -0.049 |
| SPAG9 | 0.161 | bscl2l | -0.049 |
| megf10 | 0.159 | krt18b | -0.048 |
| tyr | 0.158 | gpnmb | -0.047 |
| hsd20b2 | 0.157 | apoda.1 | -0.047 |
| slc24a5 | 0.155 | si:dkey-56m19.5 | -0.047 |
| bves | 0.155 | unm-sa821 | -0.047 |
| birc7 | 0.153 | TMEM19 | -0.047 |
| mc3r | 0.153 | pax7a | -0.046 |
| zgc:91968 | 0.152 | defbl1 | -0.046 |
| dct | 0.152 | gmps | -0.046 |
| lamp1a | 0.150 | cx30.3 | -0.046 |
| si:ch73-389b16.1 | 0.150 | lypc | -0.046 |
| CABZ01101813.1 | 0.150 | shmt1 | -0.045 |