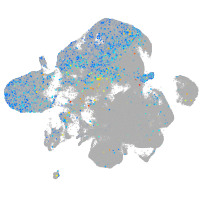
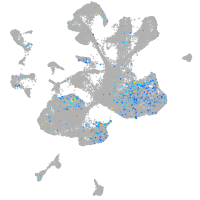
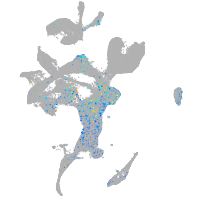
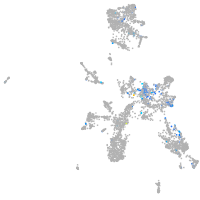
centromere protein I
ZFIN 
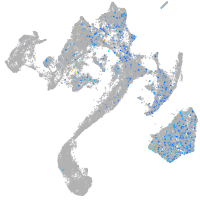
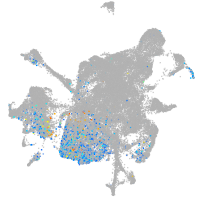
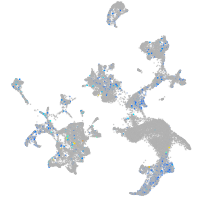
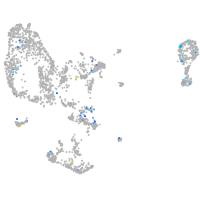
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.268 | rpl37 | -0.148 |
| plk1 | 0.252 | rps10 | -0.147 |
| cdca8 | 0.232 | zgc:114188 | -0.139 |
| tpx2 | 0.231 | zgc:92744 | -0.130 |
| mad2l1 | 0.231 | gamt | -0.128 |
| ccnb1 | 0.223 | rps17 | -0.127 |
| hmgb2a | 0.223 | gapdh | -0.112 |
| aurkb | 0.221 | gatm | -0.110 |
| dlgap5 | 0.219 | ahcy | -0.106 |
| birc5a | 0.216 | bhmt | -0.106 |
| ube2c | 0.215 | nupr1b | -0.105 |
| nusap1 | 0.214 | gpx4a | -0.105 |
| anp32a | 0.213 | atp5if1b | -0.104 |
| kifc1 | 0.212 | tpi1b | -0.101 |
| hmga1a | 0.211 | pklr | -0.099 |
| anp32b | 0.211 | mat1a | -0.099 |
| ccnb3 | 0.211 | nme2b.1 | -0.098 |
| si:ch211-69g19.2 | 0.210 | pnp4b | -0.097 |
| prc1a | 0.210 | aldob | -0.097 |
| hmgb2b | 0.209 | sod1 | -0.097 |
| cdk1 | 0.209 | apoa1b | -0.096 |
| hnrnpa1a | 0.209 | fbp1b | -0.096 |
| nucks1a | 0.208 | eno3 | -0.095 |
| seta | 0.208 | dap | -0.095 |
| dek | 0.208 | agxtb | -0.093 |
| hnrnpa1b | 0.207 | aqp12 | -0.093 |
| kpna2 | 0.204 | apoa2 | -0.092 |
| top2a | 0.203 | suclg1 | -0.092 |
| top1l | 0.202 | glud1b | -0.091 |
| aurka | 0.201 | apoa4b.1 | -0.091 |
| banf1 | 0.201 | ckba | -0.091 |
| lig1 | 0.200 | pgm1 | -0.090 |
| syncrip | 0.200 | prdx2 | -0.089 |
| zgc:110425 | 0.198 | pgk1 | -0.089 |
| ccna2 | 0.198 | mdh1aa | -0.087 |