cadherin EGF LAG seven-pass G-type receptor 1b
ZFIN 
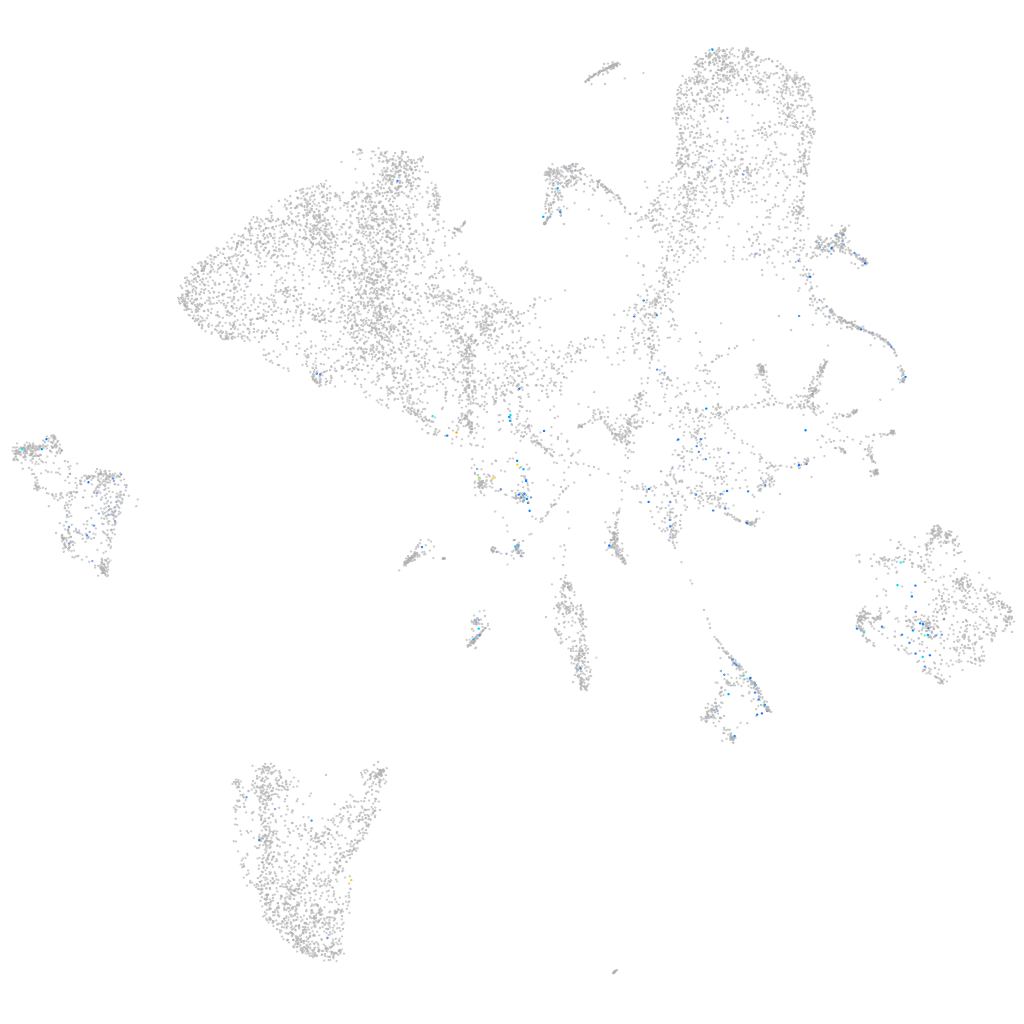
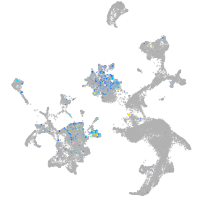
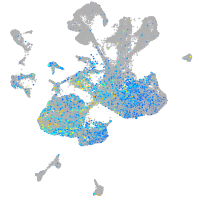
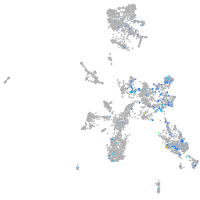
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR848840.1 | 0.199 | gamt | -0.107 |
| CABZ01087091.1 | 0.194 | gapdh | -0.106 |
| hspb15 | 0.180 | gatm | -0.090 |
| XLOC-043761 | 0.176 | apoc2 | -0.088 |
| LOC103909610 | 0.174 | mat1a | -0.087 |
| CABZ01118575.1 | 0.167 | gpx4a | -0.086 |
| CU896691.1 | 0.167 | eno3 | -0.086 |
| ankrd6a | 0.166 | apoa4b.1 | -0.085 |
| cadm3 | 0.155 | fbp1b | -0.085 |
| CU929294.1 | 0.151 | aldob | -0.085 |
| LOC101884740 | 0.151 | ahcy | -0.084 |
| LOC110439939 | 0.150 | dap | -0.082 |
| LOC101886581 | 0.145 | scp2a | -0.082 |
| XLOC-019975 | 0.144 | gcshb | -0.082 |
| FP101883.1 | 0.142 | suclg1 | -0.081 |
| CABZ01080006.1 | 0.137 | glud1b | -0.081 |
| BX649328.1 | 0.136 | gstt1a | -0.081 |
| XLOC-025051 | 0.135 | atp5if1b | -0.079 |
| sgsm1b | 0.133 | pklr | -0.079 |
| si:ch211-266k22.6 | 0.132 | bhmt | -0.078 |
| XLOC-044186 | 0.132 | agxtb | -0.078 |
| LOC108192008 | 0.130 | nipsnap3a | -0.077 |
| rx2 | 0.129 | pgm1 | -0.077 |
| otx5 | 0.128 | apoa1b | -0.077 |
| si:dkeyp-9d4.2 | 0.127 | abat | -0.076 |
| CT573366.1 | 0.126 | grhprb | -0.075 |
| fabp7a | 0.126 | suclg2 | -0.075 |
| LOC108192030 | 0.124 | msrb2 | -0.075 |
| mdka | 0.123 | srd5a2a | -0.075 |
| XLOC-016325 | 0.123 | gpd1b | -0.075 |
| si:ch211-276c2.2 | 0.122 | aldh7a1 | -0.074 |
| atp1a1b | 0.119 | aqp12 | -0.074 |
| lrrc3 | 0.118 | ttc36 | -0.074 |
| msna | 0.118 | pgk1 | -0.074 |
| bcam | 0.118 | slco1d1 | -0.074 |