cerebellar degeneration-related protein 2-like
ZFIN 
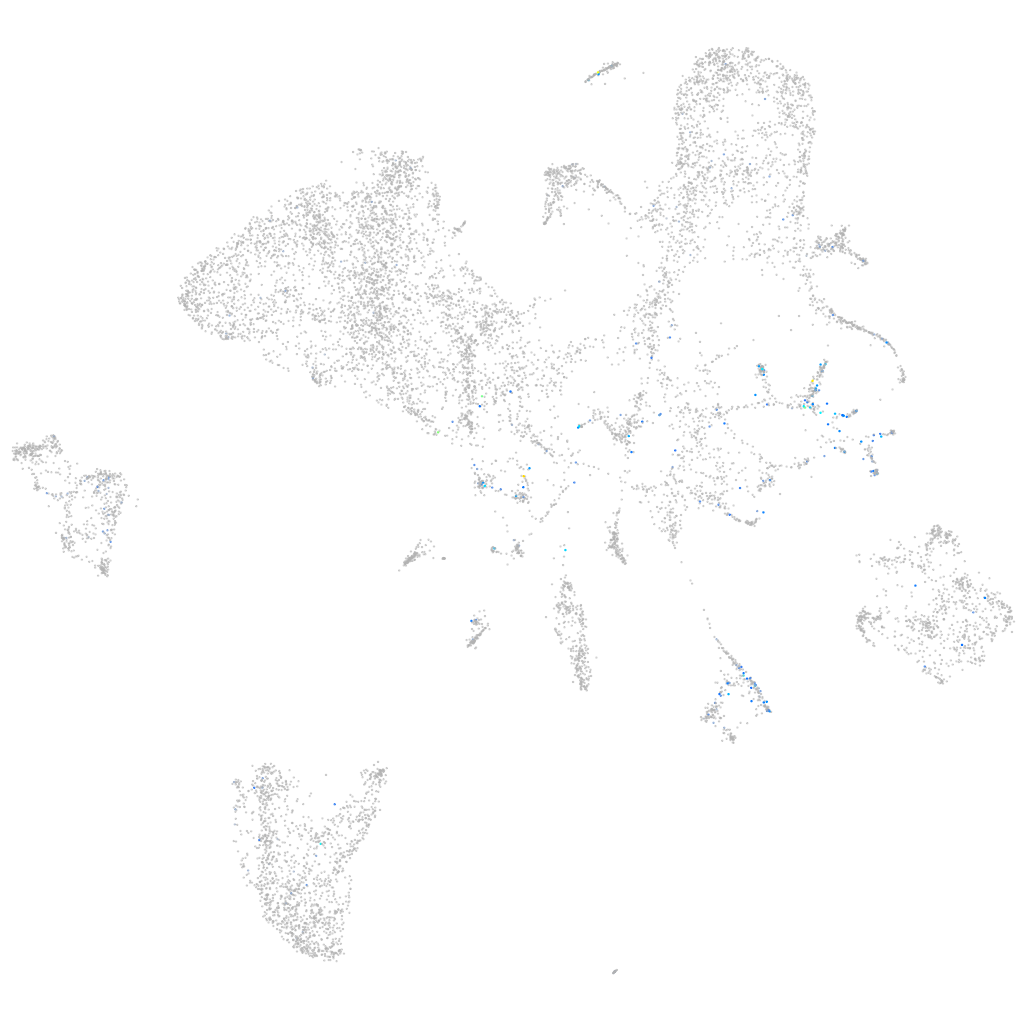
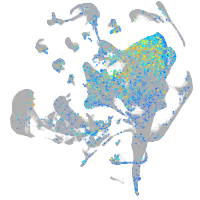
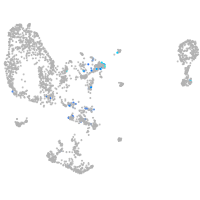
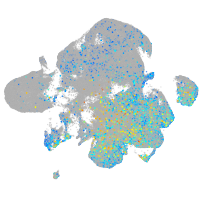
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.206 | gapdh | -0.108 |
| neurod1 | 0.190 | gamt | -0.102 |
| vat1 | 0.188 | aldob | -0.099 |
| scg3 | 0.185 | gatm | -0.092 |
| rasd4 | 0.182 | fbp1b | -0.091 |
| pax6b | 0.181 | ahcy | -0.085 |
| kcnj11 | 0.178 | mat1a | -0.085 |
| LOC100537384 | 0.173 | apoa4b.1 | -0.085 |
| gpr142 | 0.170 | apoc2 | -0.084 |
| BX927329.2 | 0.169 | bhmt | -0.084 |
| arxa | 0.169 | gstt1a | -0.082 |
| tspan7b | 0.165 | apoa1b | -0.080 |
| BX546499.2 | 0.163 | aldh6a1 | -0.080 |
| syt1a | 0.163 | dap | -0.080 |
| si:dkey-153k10.9 | 0.160 | scp2a | -0.080 |
| gapdhs | 0.159 | sult2st2 | -0.078 |
| si:ch73-287m6.1 | 0.159 | afp4 | -0.078 |
| vamp2 | 0.159 | gpx4a | -0.077 |
| rnasekb | 0.157 | glud1b | -0.077 |
| rprmb | 0.156 | agxtb | -0.076 |
| egr4 | 0.155 | suclg2 | -0.076 |
| insm1b | 0.155 | agxta | -0.076 |
| pcsk1nl | 0.154 | nipsnap3a | -0.076 |
| nkx2.2a | 0.154 | pnp4b | -0.076 |
| lysmd2 | 0.154 | grhprb | -0.075 |
| insm1a | 0.153 | rdh1 | -0.075 |
| tox | 0.152 | gnmt | -0.075 |
| tango2 | 0.152 | g6pca.2 | -0.074 |
| mir7a-1 | 0.151 | mgst1.2 | -0.074 |
| jpt1b | 0.150 | aldh7a1 | -0.074 |
| c2cd4a | 0.149 | aqp12 | -0.073 |
| CR812832.1 | 0.147 | msrb2 | -0.073 |
| tenm1 | 0.146 | cx32.3 | -0.073 |
| pcsk2 | 0.145 | haao | -0.073 |
| capsla | 0.145 | ftcd | -0.073 |