cyclin-dependent kinase 2 associated protein 2
ZFIN 
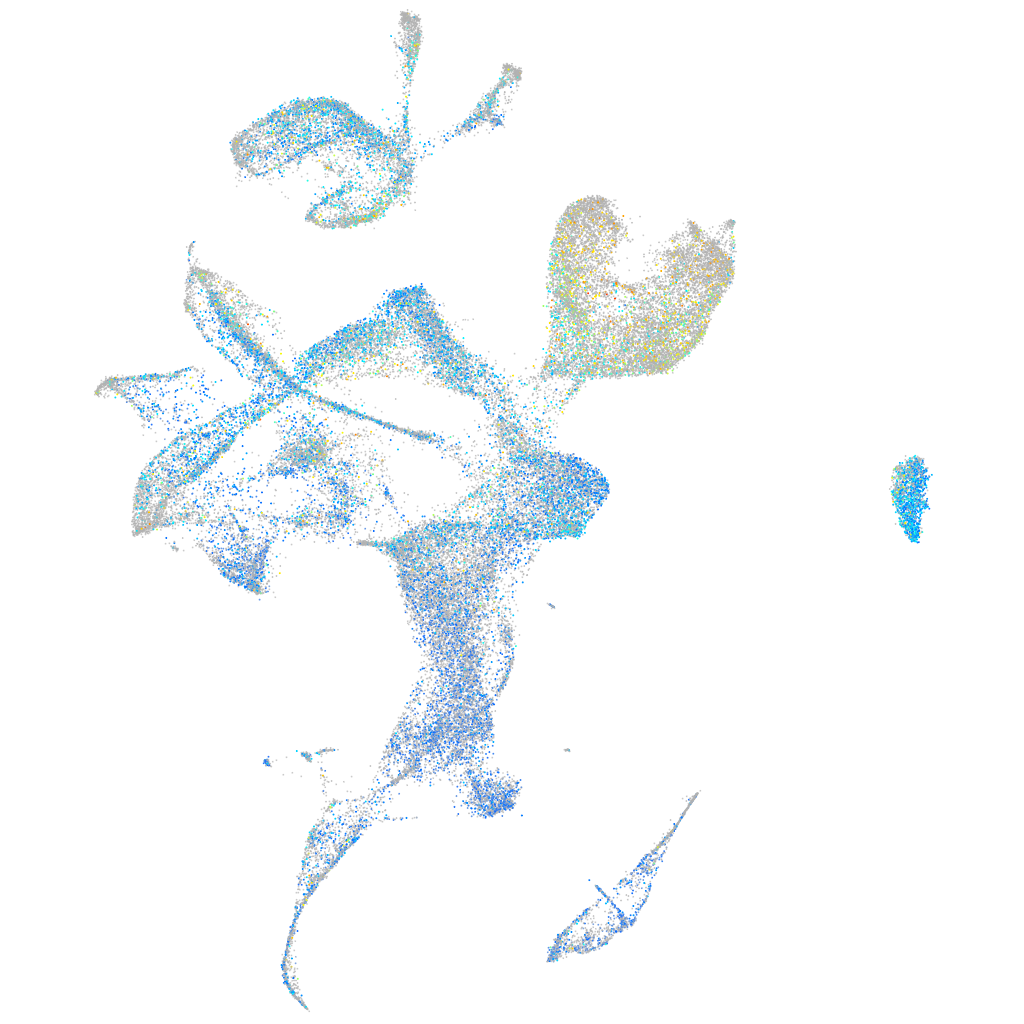
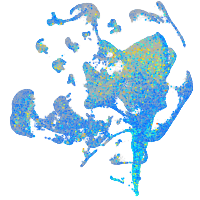
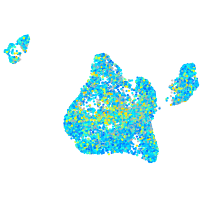
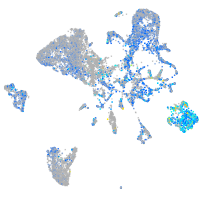
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.110 | CR383676.1 | -0.063 |
| hesx1 | 0.106 | si:dkey-16p21.8 | -0.061 |
| hnrnpaba | 0.106 | bhlhe40 | -0.060 |
| asb11 | 0.105 | pvalb2 | -0.058 |
| shisa2a | 0.104 | pvalb1 | -0.057 |
| cyp26a1 | 0.102 | sncgb | -0.056 |
| tdgf1 | 0.101 | apoa2 | -0.055 |
| stm | 0.100 | eno1a | -0.054 |
| hnrnpa0b | 0.100 | FQ323156.1 | -0.053 |
| hnrnpabb | 0.099 | fzd5 | -0.053 |
| nnr | 0.099 | actc1b | -0.052 |
| s100a1 | 0.098 | her12 | -0.052 |
| COX7A2 | 0.098 | vsx2 | -0.051 |
| khdrbs1a | 0.097 | her4.1 | -0.049 |
| si:ch211-152c2.3 | 0.097 | apoa1b | -0.048 |
| apoeb | 0.097 | glula | -0.047 |
| sec61b | 0.097 | her4.2 | -0.046 |
| cirbpb | 0.096 | XLOC-003690 | -0.046 |
| polr3gla | 0.096 | her15.1 | -0.045 |
| LOC108190024 | 0.096 | CELA1 (1 of many) | -0.045 |
| sox19a | 0.096 | aldocb | -0.044 |
| pkdccb | 0.095 | notch1b | -0.043 |
| foxd5 | 0.094 | rgs8 | -0.042 |
| syncrip | 0.093 | prss1 | -0.042 |
| NC-002333.4 | 0.093 | prss59.2 | -0.042 |
| ilf3b | 0.092 | zgc:158463 | -0.041 |
| si:ch1073-80i24.3 | 0.091 | rrm2 | -0.041 |
| rbm4.3 | 0.091 | zgc:56493 | -0.041 |
| nr6a1a | 0.090 | dut | -0.041 |
| ilf2 | 0.090 | si:ch73-256g18.2 | -0.041 |
| srsf3b | 0.088 | rgs16 | -0.041 |
| zmp:0000000624 | 0.088 | prss59.1 | -0.040 |
| hnrnpa0a | 0.088 | XLOC-003692 | -0.040 |
| alcamb | 0.088 | COX7A2 (1 of many) | -0.040 |
| hnrnpa1a | 0.087 | tnni2a.4 | -0.039 |