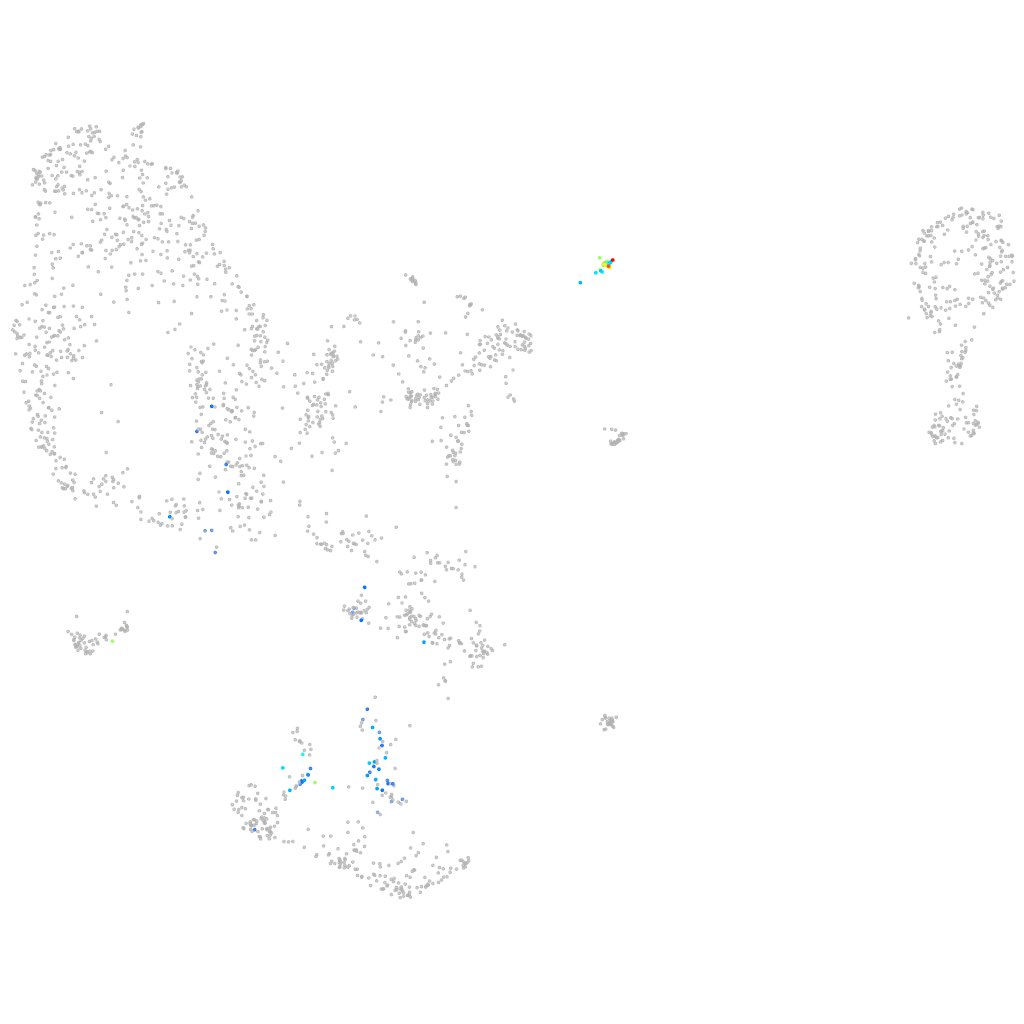
"cadherin 16, KSP-cadherin"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| frzb | 0.763 | aldob | -0.159 |
| stc1l | 0.705 | gapdh | -0.135 |
| inka1a | 0.700 | gstt1a | -0.131 |
| pcsk2 | 0.680 | grhprb | -0.128 |
| ngb | 0.624 | si:dkey-194e6.1 | -0.125 |
| npdc1a | 0.586 | upb1 | -0.123 |
| si:ch211-163l21.8 | 0.574 | ahcy | -0.122 |
| lin7a | 0.571 | pdzk1ip1 | -0.122 |
| slc41a1 | 0.565 | cox6a2 | -0.120 |
| slc8a4b | 0.548 | slc26a1 | -0.118 |
| slc43a2a | 0.544 | acy3.2 | -0.117 |
| prex2 | 0.522 | dab2 | -0.117 |
| anos1a | 0.516 | fbp1b | -0.116 |
| apcdd1l | 0.507 | pnp6 | -0.115 |
| hhla2a.2 | 0.501 | cubn | -0.115 |
| areg | 0.492 | lrp2a | -0.114 |
| fgf23 | 0.488 | si:dkey-33i11.9 | -0.113 |
| tgfa | 0.486 | dpys | -0.113 |
| cnnm2b | 0.480 | alpl | -0.112 |
| pip5k1ca | 0.463 | npm1a | -0.111 |
| CT030153.1 | 0.462 | phyhd1 | -0.109 |
| tnfsf12 | 0.448 | slc9a3r1a | -0.109 |
| vwa5b2 | 0.445 | gpt2l | -0.108 |
| zgc:195023 | 0.442 | nit2 | -0.108 |
| sall1a | 0.441 | eps8l3b | -0.107 |
| errfi1a | 0.432 | gpd1b | -0.107 |
| zgc:112332 | 0.429 | ftcd | -0.107 |
| grin2cb | 0.427 | hao1 | -0.105 |
| FQ790211.1 | 0.427 | eps8l3a | -0.104 |
| si:ch211-247j9.1 | 0.422 | aqp8b | -0.102 |
| cnih2 | 0.421 | gcshb | -0.102 |
| CABZ01073166.1 | 0.419 | adka | -0.102 |
| wif1 | 0.412 | gnpda1 | -0.102 |
| admb | 0.410 | hoga1 | -0.102 |
| pln | 0.408 | dpydb | -0.102 |