CDC42 effector protein (Rho GTPase binding) 5
ZFIN 
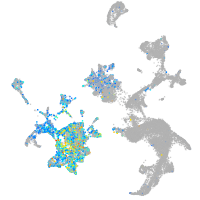
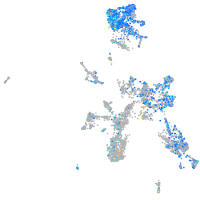
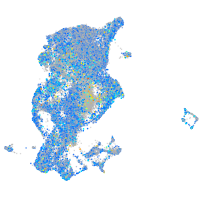
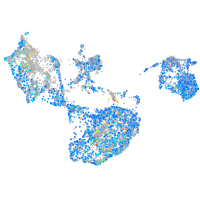
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.156 | mia | -0.067 |
| pmp22a | 0.135 | matn1 | -0.064 |
| actb1 | 0.132 | fgfbp2b | -0.063 |
| msna | 0.129 | snorc | -0.059 |
| zgc:153867 | 0.129 | gpm6aa | -0.059 |
| si:ch211-286o17.1 | 0.128 | col11a2 | -0.058 |
| si:dkey-261h17.1 | 0.125 | stmn1b | -0.057 |
| mfap2 | 0.125 | tnxba | -0.057 |
| actb2 | 0.118 | ckbb | -0.055 |
| cnn2 | 0.117 | apoa2 | -0.055 |
| myl12.1 | 0.117 | ucmab | -0.054 |
| sept2 | 0.115 | wisp3 | -0.054 |
| gng5 | 0.115 | apoa1b | -0.053 |
| marcksl1a | 0.112 | si:dkey-151g10.6 | -0.053 |
| akap12b | 0.108 | epyc | -0.051 |
| sept10 | 0.108 | acana | -0.051 |
| flot2a | 0.105 | elavl3 | -0.051 |
| sri | 0.104 | rpl38 | -0.051 |
| klf6a | 0.104 | ptgdsa | -0.050 |
| rac1a | 0.102 | otos | -0.048 |
| tpm3 | 0.102 | pvalb2 | -0.047 |
| cfl1 | 0.101 | rtn1a | -0.047 |
| cdc42l | 0.100 | sncb | -0.046 |
| tuba8l3 | 0.099 | pcolceb | -0.046 |
| krt18b | 0.099 | VIT | -0.045 |
| tuba8l4 | 0.099 | CU467822.1 | -0.045 |
| marcksl1b | 0.098 | rps29 | -0.045 |
| hmgb2a | 0.098 | rbp4 | -0.045 |
| si:dkey-238c7.16 | 0.097 | tmsb | -0.044 |
| arhgdia | 0.097 | pvalb1 | -0.044 |
| colec12 | 0.096 | rpl39 | -0.044 |
| hmgn2 | 0.095 | csgalnact1a | -0.044 |
| tubb4b | 0.094 | col2a1a | -0.044 |
| ywhaz | 0.094 | col9a1a | -0.043 |
| rhoab | 0.094 | LOC100330861 | -0.043 |