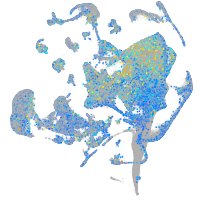
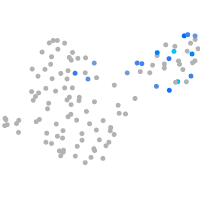
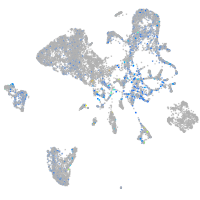
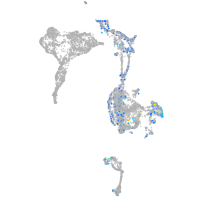
cyclin T1
ZFIN 
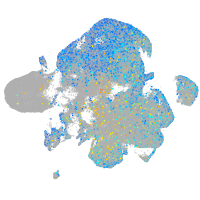
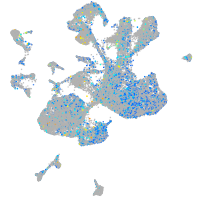
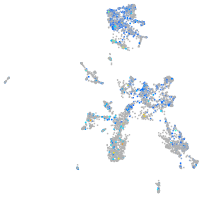
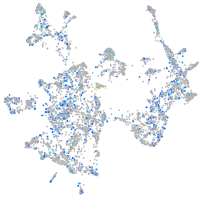
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.108 | glula | -0.054 |
| tpx2 | 0.101 | cx43 | -0.054 |
| cenpf | 0.099 | dap1b | -0.051 |
| top2a | 0.098 | slc3a2a | -0.051 |
| plk1 | 0.094 | acbd7 | -0.050 |
| seta | 0.093 | cox4i2 | -0.050 |
| ccna2 | 0.093 | slc4a4a | -0.049 |
| ccnb1 | 0.093 | efhd1 | -0.049 |
| kif11 | 0.092 | ppdpfb | -0.048 |
| dlgap5 | 0.091 | slc1a2b | -0.048 |
| cdc20 | 0.090 | ptn | -0.048 |
| hmga1a | 0.088 | anxa13 | -0.047 |
| si:ch211-222l21.1 | 0.087 | apoa2 | -0.047 |
| aspm | 0.087 | mt2 | -0.047 |
| zgc:165555.3 | 0.087 | atp1a1b | -0.046 |
| ube2c | 0.087 | gapdhs | -0.045 |
| cirbpa | 0.087 | fabp7a | -0.045 |
| kifc1 | 0.087 | si:ch211-66e2.5 | -0.045 |
| kpna2 | 0.086 | zgc:153704 | -0.045 |
| nusap1 | 0.086 | ndrg3a | -0.045 |
| g2e3 | 0.086 | ppap2d | -0.044 |
| lbr | 0.086 | ptgdsb.2 | -0.043 |
| tacc3 | 0.085 | cebpd | -0.043 |
| cenpe | 0.085 | hepacama | -0.043 |
| aurkb | 0.085 | cyp2ad3 | -0.043 |
| si:ch211-288g17.3 | 0.085 | smox | -0.042 |
| aurka | 0.084 | pvalb1 | -0.042 |
| khdrbs1a | 0.084 | nupr1a | -0.041 |
| ptmab | 0.084 | cyp3c1 | -0.041 |
| cdk1 | 0.084 | ckbb | -0.041 |
| hmgb2a | 0.084 | pvalb2 | -0.041 |
| hnrnpaba | 0.084 | mylpfa | -0.040 |
| si:ch211-69g19.2 | 0.084 | cotl1 | -0.040 |
| h3f3a | 0.084 | lpin1 | -0.040 |
| cdca8 | 0.084 | apoa1b | -0.039 |