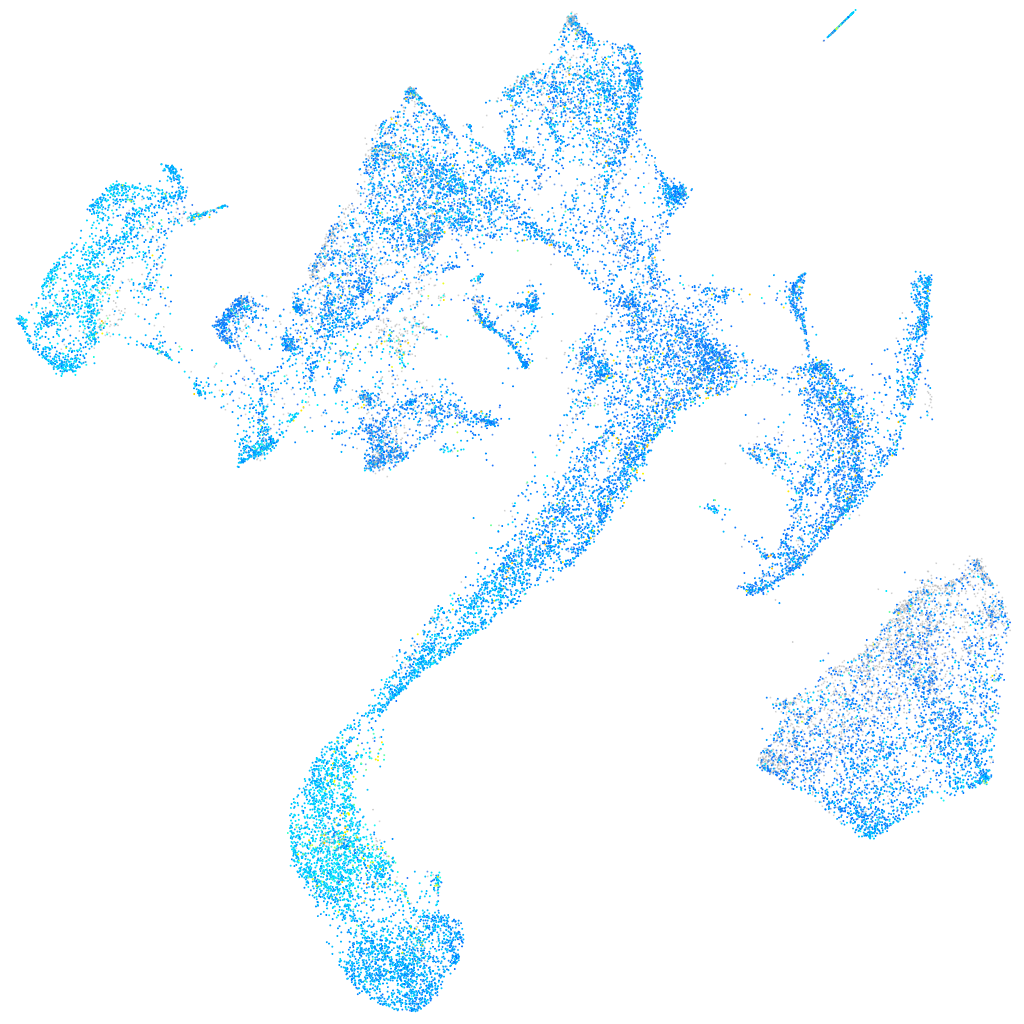
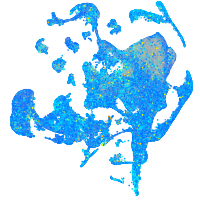
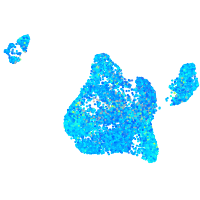
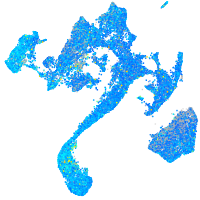
cyclin G1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldoab | 0.393 | pabpc1a | -0.360 |
| tmem38a | 0.385 | si:ch73-1a9.3 | -0.354 |
| ckmb | 0.384 | ptmab | -0.350 |
| gapdh | 0.380 | si:ch211-222l21.1 | -0.319 |
| ak1 | 0.379 | hsp90ab1 | -0.315 |
| ckma | 0.379 | cirbpa | -0.305 |
| actc1b | 0.378 | hmgb2a | -0.289 |
| acta1b | 0.374 | hmgb2b | -0.288 |
| neb | 0.370 | anp32a | -0.285 |
| atp2a1 | 0.370 | acin1a | -0.283 |
| actn3b | 0.370 | h3f3d | -0.278 |
| eno3 | 0.369 | nucks1a | -0.272 |
| ldb3b | 0.367 | hnrnpaba | -0.271 |
| tnnc2 | 0.367 | seta | -0.270 |
| actn3a | 0.367 | si:ch73-281n10.2 | -0.269 |
| srl | 0.366 | hmga1a | -0.267 |
| CABZ01078594.1 | 0.365 | syncrip | -0.267 |
| ldb3a | 0.362 | NC-002333.4 | -0.266 |
| ttn.2 | 0.362 | ncl | -0.266 |
| mybphb | 0.361 | hnrnpa1a | -0.264 |
| casq2 | 0.361 | marcksb | -0.262 |
| tnnt3a | 0.360 | hmgn7 | -0.260 |
| myom1a | 0.359 | hmgn2 | -0.258 |
| CABZ01072309.1 | 0.359 | ppiaa | -0.253 |
| smyd1a | 0.358 | hmgn6 | -0.252 |
| pgam2 | 0.357 | cirbpb | -0.252 |
| ttn.1 | 0.356 | hnrnpa1b | -0.251 |
| ank1a | 0.355 | eif5a2 | -0.247 |
| tmod4 | 0.354 | cx43.4 | -0.244 |
| hhatla | 0.354 | cdx4 | -0.244 |
| suclg1 | 0.354 | khdrbs1a | -0.244 |
| eno1a | 0.353 | h2afvb | -0.242 |
| tpma | 0.353 | nop58 | -0.242 |
| si:ch73-367p23.2 | 0.352 | rps18 | -0.242 |
| cav3 | 0.352 | anp32b | -0.241 |