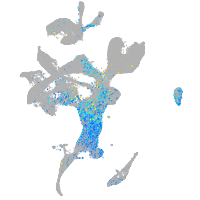
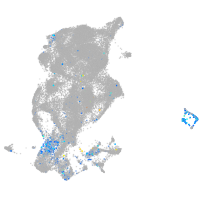
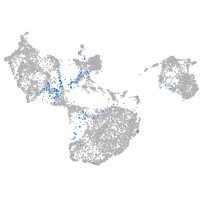
cyclin E2
ZFIN 
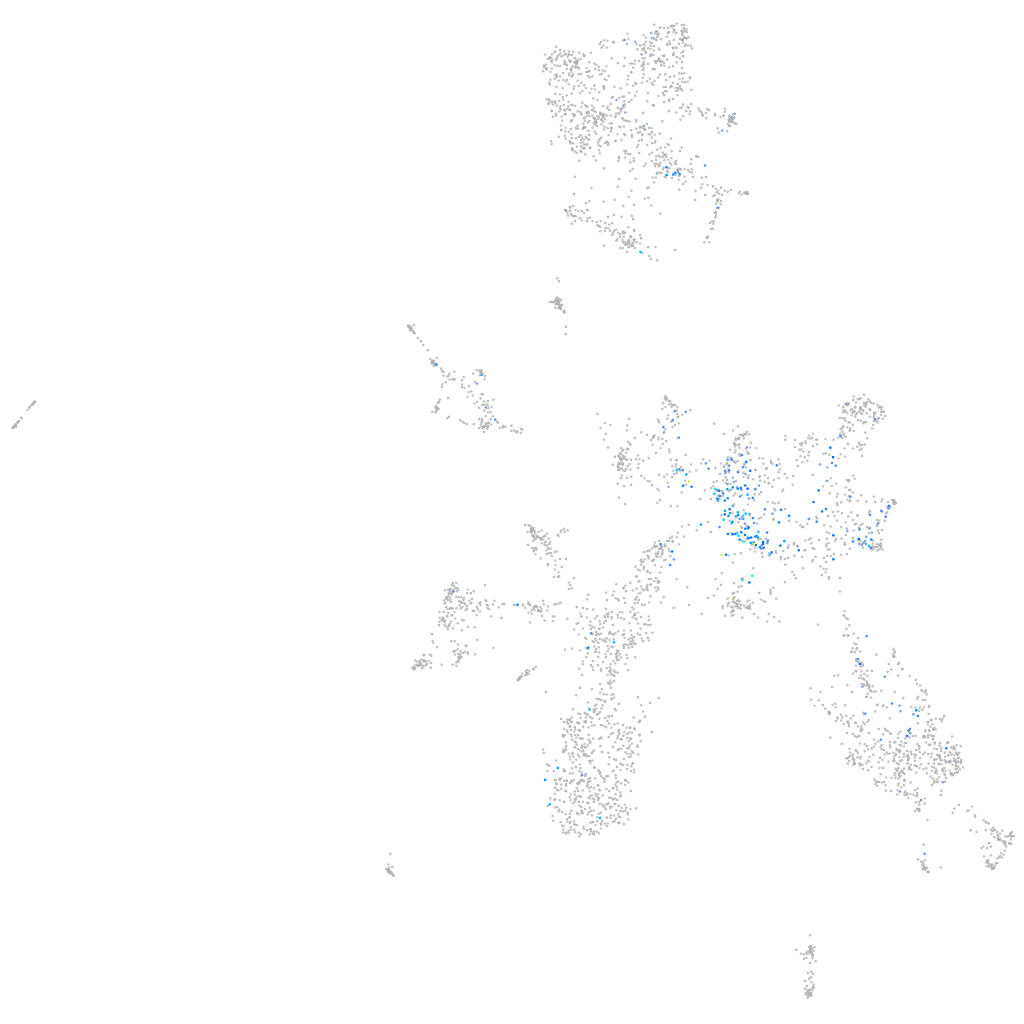
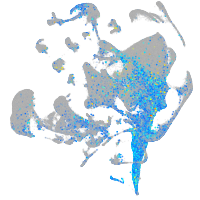
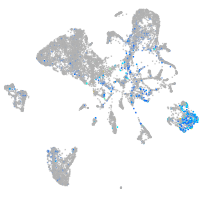
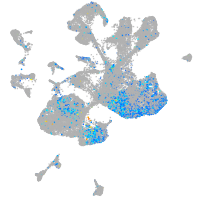
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slbp | 0.490 | txn | -0.210 |
| zgc:110540 | 0.462 | gstp1 | -0.188 |
| tk1 | 0.442 | selenow1 | -0.185 |
| esco2 | 0.438 | anxa5b | -0.184 |
| CABZ01005379.1 | 0.425 | gapdhs | -0.170 |
| rrm2 | 0.414 | tmem59 | -0.168 |
| cdc6 | 0.409 | cldnh | -0.166 |
| nasp | 0.406 | bik | -0.165 |
| pcna | 0.390 | selenow2b | -0.159 |
| asf1ba | 0.387 | calm1a | -0.155 |
| rrm1 | 0.385 | CR383676.1 | -0.153 |
| chaf1a | 0.381 | wu:fb18f06 | -0.151 |
| dut | 0.377 | nfe2l2a | -0.151 |
| rpa2 | 0.368 | ccni | -0.147 |
| stmn1a | 0.361 | atp1b1a | -0.145 |
| fbxo5 | 0.359 | fxyd1 | -0.144 |
| exo1 | 0.358 | spint2 | -0.140 |
| mibp | 0.358 | smdt1b | -0.137 |
| LOC100330864 | 0.357 | ppdpfb | -0.137 |
| fen1 | 0.352 | calm1b | -0.137 |
| lig1 | 0.351 | gabarapa | -0.136 |
| mcm7 | 0.349 | vamp2 | -0.135 |
| her2 | 0.346 | h1f0 | -0.134 |
| rfc4 | 0.345 | tprg1 | -0.134 |
| mcm4 | 0.343 | gsto2 | -0.133 |
| dck | 0.337 | gbgt1l4 | -0.132 |
| prim2 | 0.336 | gnb1a | -0.131 |
| rpa3 | 0.335 | calb2a | -0.131 |
| atad5a | 0.332 | atp6v1g1 | -0.130 |
| dek | 0.323 | chga | -0.130 |
| chek1 | 0.321 | si:ch1073-443f11.2 | -0.130 |
| asf1bb | 0.319 | ponzr6 | -0.130 |
| pola2 | 0.319 | tnks1bp1 | -0.129 |
| si:ch211-156b7.4 | 0.311 | ip6k2a | -0.127 |
| rbbp4 | 0.311 | si:ch211-153b23.5 | -0.127 |