chemokine (C-C motif) ligand 20b
ZFIN 
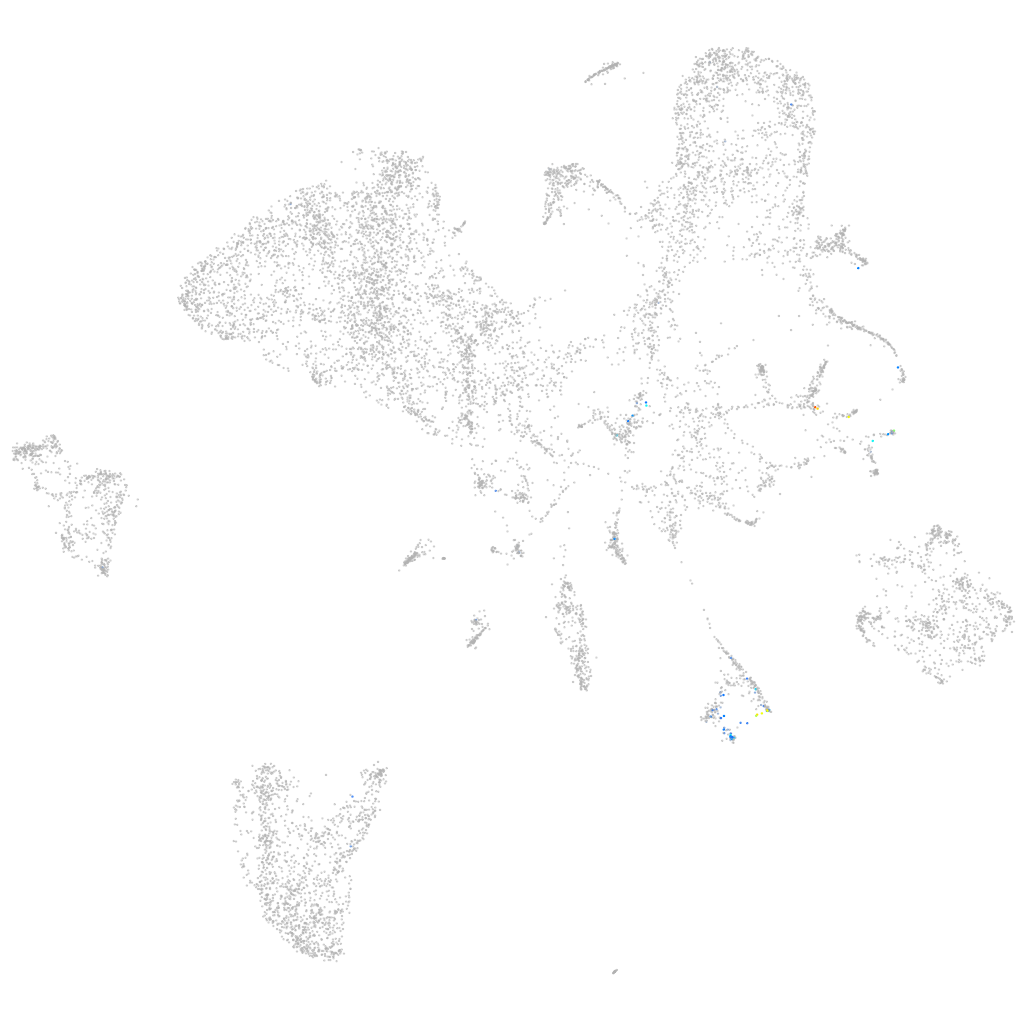
Other cell groups


all cells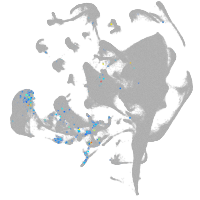 |
blastomeres |
PGCs |
endoderm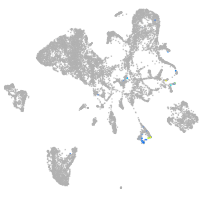 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 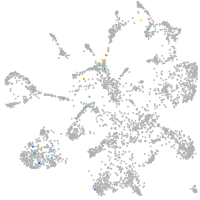 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 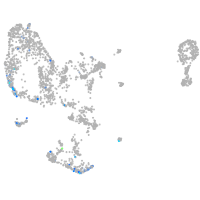 |
neural |
spinal cord / glia |
eye |
taste / olfactory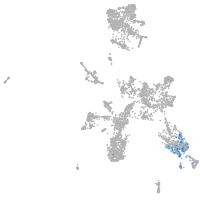 |
otic / lateral line |
epidermis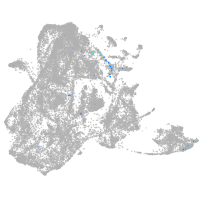 |
periderm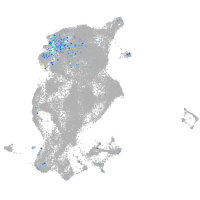 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL935044.4 | 0.352 | gapdh | -0.070 |
| si:ch211-217k17.10 | 0.286 | gatm | -0.061 |
| LOC103910220 | 0.259 | fabp3 | -0.059 |
| timp2b | 0.240 | fbp1b | -0.058 |
| si:dkey-192g7.3 | 0.239 | bhmt | -0.057 |
| runx2a | 0.235 | ahcy | -0.057 |
| cd151 | 0.231 | gamt | -0.055 |
| CR855337.1 | 0.230 | gpx4a | -0.055 |
| mxra8b | 0.227 | mat1a | -0.054 |
| CABZ01055365.1 | 0.223 | apoc2 | -0.053 |
| si:ch73-329n5.1 | 0.221 | aldob | -0.053 |
| amotl2b | 0.218 | hspe1 | -0.053 |
| pnp4a | 0.218 | scp2a | -0.052 |
| loxl5a | 0.213 | aqp12 | -0.052 |
| CR762483.1 | 0.211 | ssr3 | -0.050 |
| pimr51 | 0.210 | gstr | -0.050 |
| sim2 | 0.209 | afp4 | -0.050 |
| saa | 0.203 | gcshb | -0.050 |
| f3a | 0.203 | apoa4b.1 | -0.050 |
| ccl20a.3 | 0.202 | gstt1a | -0.049 |
| gna15.1 | 0.202 | apoa1b | -0.049 |
| cxcl8a | 0.199 | si:dkey-16p21.8 | -0.049 |
| sim1b | 0.185 | sult2st2 | -0.048 |
| gip | 0.182 | apoc1 | -0.048 |
| ctgfa | 0.179 | aldh6a1 | -0.048 |
| si:dkey-15b23.3 | 0.178 | g6pca.2 | -0.048 |
| slc6a14 | 0.176 | grhprb | -0.047 |
| anxa3b | 0.175 | slc37a4a | -0.047 |
| BX908782.2 | 0.173 | atp5mc1 | -0.047 |
| snap23.2 | 0.172 | agxta | -0.046 |
| cyr61 | 0.171 | dhrs9 | -0.046 |
| spock3 | 0.171 | mdh1aa | -0.046 |
| sftpba | 0.170 | suclg2 | -0.046 |
| cygb1 | 0.169 | dpydb | -0.046 |
| BX571707.1 | 0.169 | abat | -0.046 |