coiled-coil domain containing 39
ZFIN 
Other cell groups


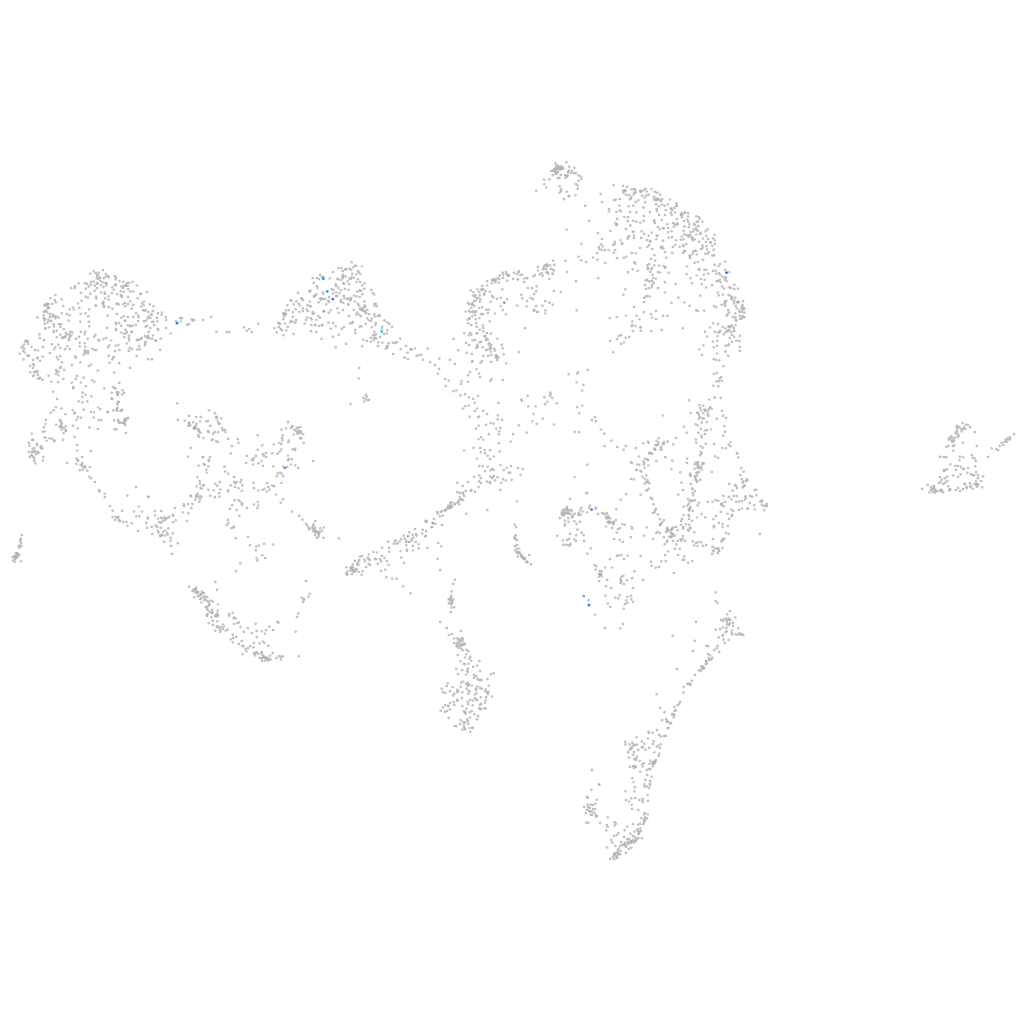
all cells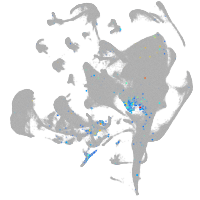 |
blastomeres |
PGCs |
endoderm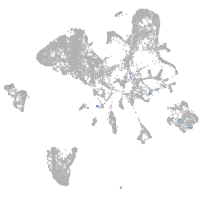 |
axial mesoderm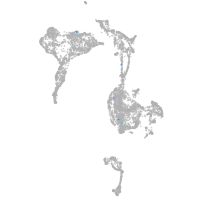 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia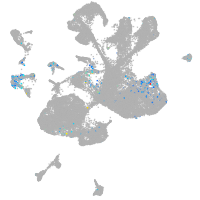 |
eye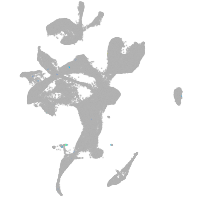 |
taste / olfactory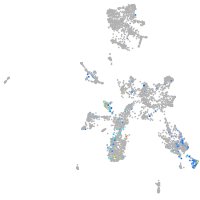 |
otic / lateral line |
epidermis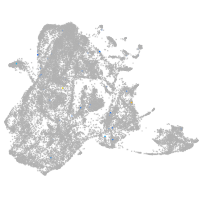 |
periderm |
fin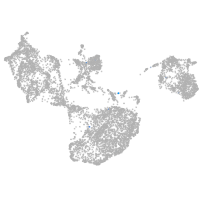 |
ionocytes / mucous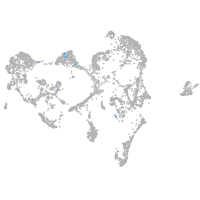 |
pigment cells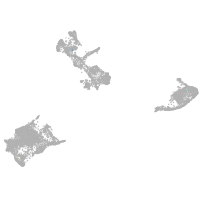 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| btg4 | 0.374 | hypk | -0.039 |
| es1 | 0.374 | h2afvb | -0.036 |
| ttc34 | 0.374 | fam32a | -0.035 |
| tekt2 | 0.374 | ywhabl | -0.034 |
| kcnh1a | 0.344 | drap1 | -0.033 |
| si:dkey-267j14.7 | 0.341 | si:ch73-234b20.5 | -0.033 |
| CR392042.1 | 0.325 | tma7 | -0.033 |
| dnah3 | 0.319 | alyref | -0.032 |
| si:ch1073-349o24.2 | 0.318 | eif2s2 | -0.031 |
| zgc:195356 | 0.307 | znf593 | -0.030 |
| dnah5l | 0.305 | cirbpa | -0.030 |
| spaca9 | 0.304 | blcap | -0.030 |
| crocc2 | 0.304 | CABZ01068251.1 | -0.030 |
| tbata | 0.298 | si:dkey-151g10.6 | -0.030 |
| si:ch73-174h16.5 | 0.293 | znf185 | -0.030 |
| CU929451.2 | 0.292 | snrpd3l | -0.030 |
| mthfd1a | 0.282 | glrx5 | -0.029 |
| CR407547.1 | 0.281 | srsf6b | -0.029 |
| zgc:92275 | 0.280 | si:ch211-51e12.7 | -0.028 |
| armc3 | 0.277 | tmed9 | -0.028 |
| zbbx | 0.274 | eif4g1a | -0.028 |
| si:dkey-161j23.6 | 0.274 | fzr1a | -0.028 |
| kcnn4 | 0.267 | rcc2 | -0.027 |
| dnaaf2 | 0.266 | ncoa4 | -0.027 |
| foxj1a | 0.257 | nhp2 | -0.027 |
| zmynd10 | 0.257 | psma6a | -0.027 |
| cfap157 | 0.245 | snrpb | -0.027 |
| si:dkey-52j6.3 | 0.242 | lmnb2 | -0.026 |
| spata17 | 0.241 | hist2h2l | -0.026 |
| si:ch211-198k9.6 | 0.237 | fkbp4 | -0.026 |
| mycbpap | 0.229 | hacd2 | -0.026 |
| peli1a | 0.229 | si:dkey-262k9.2 | -0.026 |
| epha6 | 0.227 | znf395a | -0.026 |
| kif9 | 0.226 | snrpg | -0.026 |
| dnah1 | 0.223 | uqcc3 | -0.025 |