coiled-coil domain containing 174
ZFIN 
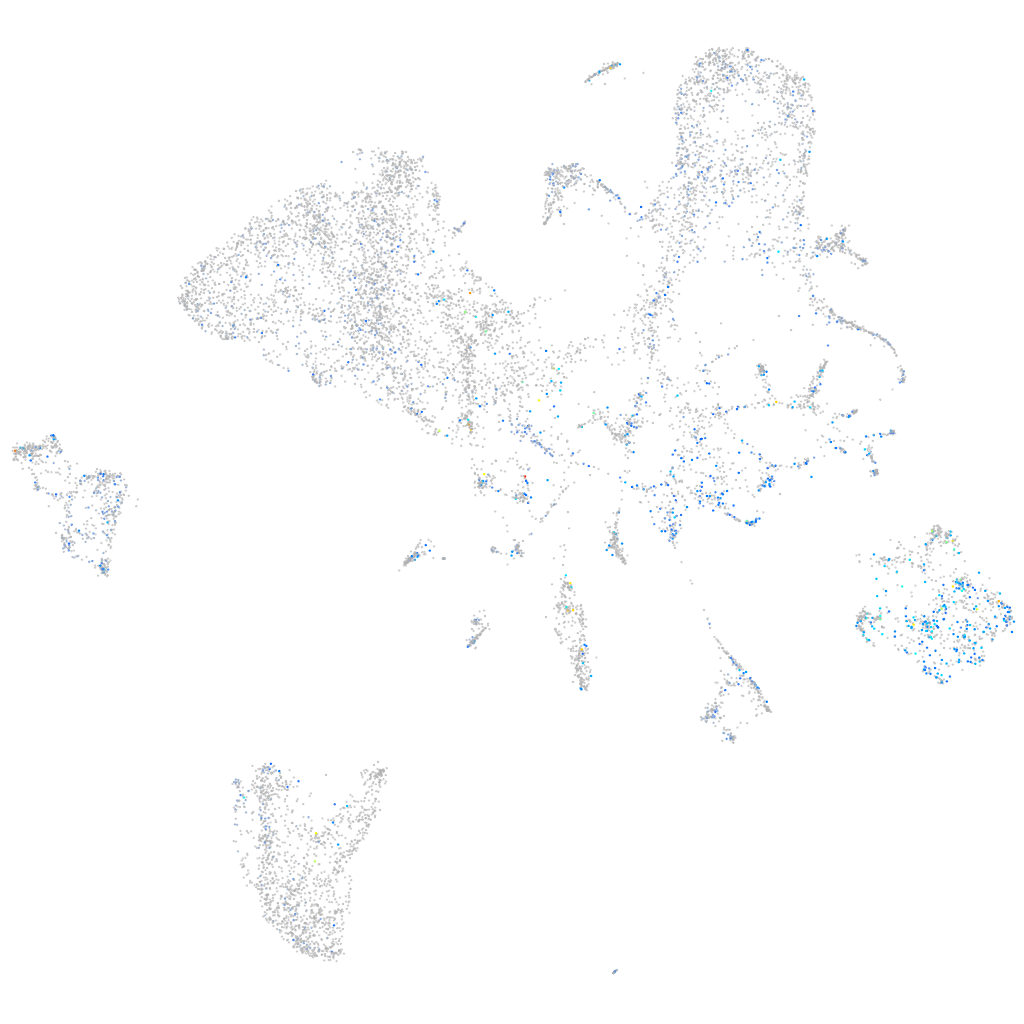
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksl1b | 0.250 | gamt | -0.173 |
| akap12b | 0.245 | ahcy | -0.171 |
| marcksb | 0.244 | gapdh | -0.169 |
| si:ch211-152c2.3 | 0.244 | rpl37 | -0.167 |
| cx43.4 | 0.235 | gatm | -0.161 |
| hspb1 | 0.234 | zgc:114188 | -0.151 |
| syncrip | 0.234 | rps10 | -0.151 |
| khdrbs1a | 0.231 | bhmt | -0.148 |
| hmga1a | 0.230 | nme2b.1 | -0.147 |
| acin1a | 0.228 | eno3 | -0.143 |
| anp32a | 0.226 | zgc:92744 | -0.143 |
| seta | 0.224 | nupr1b | -0.139 |
| hnrnpa0b | 0.223 | apoa1b | -0.138 |
| nucks1a | 0.223 | aldh7a1 | -0.137 |
| si:dkey-66i24.9 | 0.222 | cx32.3 | -0.137 |
| hnrnpa1b | 0.222 | mat1a | -0.136 |
| ilf3b | 0.221 | sod1 | -0.134 |
| hnrnpaba | 0.220 | pnp4b | -0.134 |
| hmgn6 | 0.219 | scp2a | -0.134 |
| ppig | 0.219 | agxtb | -0.134 |
| smarca4a | 0.218 | glud1b | -0.131 |
| hdac1 | 0.218 | suclg1 | -0.130 |
| zmat2 | 0.218 | apoa2 | -0.130 |
| si:ch73-1a9.3 | 0.216 | apoa4b.1 | -0.129 |
| snrpa | 0.216 | gpx4a | -0.129 |
| hnrnpub | 0.215 | abat | -0.129 |
| hnrnpabb | 0.214 | pklr | -0.129 |
| h3f3d | 0.214 | fbp1b | -0.128 |
| snrpd3l | 0.214 | dap | -0.128 |
| h2afvb | 0.214 | eef1da | -0.128 |
| cbx3a | 0.213 | rps17 | -0.127 |
| hmgb2a | 0.213 | agxta | -0.124 |
| si:ch73-281n10.2 | 0.213 | aqp12 | -0.124 |
| hnrnpa1a | 0.212 | gnmt | -0.123 |
| smc1al | 0.212 | aldh6a1 | -0.123 |