coiled-coil domain containing 13
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 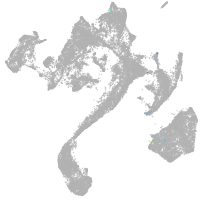 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia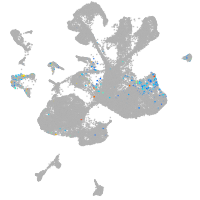 |
eye |
taste / olfactory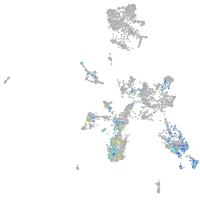 |
otic / lateral line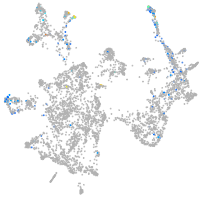 |
epidermis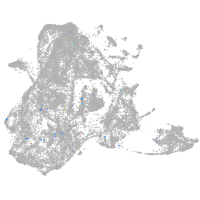 |
periderm |
fin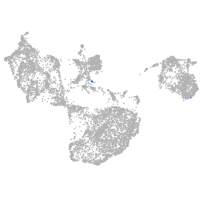 |
ionocytes / mucous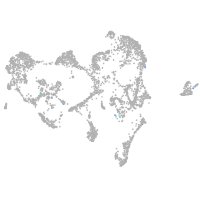 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crocc2 | 0.347 | gcshb | -0.126 |
| cfap52 | 0.332 | dab2 | -0.120 |
| tekt1 | 0.320 | si:ch211-139a5.9 | -0.119 |
| spaca9 | 0.317 | zgc:136493 | -0.113 |
| odf3l2b | 0.310 | si:dkey-16p21.8 | -0.112 |
| cdc25d | 0.297 | cryl1 | -0.106 |
| sept8b | 0.297 | pnp6 | -0.106 |
| ccdc151 | 0.297 | suclg2 | -0.106 |
| pih1d3 | 0.293 | aldob | -0.105 |
| ttc25 | 0.290 | cox6a2 | -0.105 |
| ccdc173 | 0.288 | gpt2l | -0.105 |
| casc1 | 0.286 | epdl2 | -0.104 |
| ribc2 | 0.286 | gstp1 | -0.104 |
| dnali1 | 0.283 | sod1 | -0.104 |
| gas8 | 0.282 | dpys | -0.103 |
| rsph9 | 0.282 | glud1b | -0.102 |
| efhc2 | 0.281 | ASS1 | -0.102 |
| xrra1 | 0.280 | si:dkey-33i11.9 | -0.101 |
| si:ch211-163l21.7 | 0.279 | akr7a3 | -0.101 |
| cluap1 | 0.279 | prdx2 | -0.101 |
| ift172 | 0.279 | atp2b1a | -0.099 |
| lrp2bp | 0.279 | slc22a13b | -0.099 |
| cetn4 | 0.278 | xirp1 | -0.099 |
| tekt2 | 0.276 | lgmn | -0.098 |
| mycbpap | 0.276 | amt | -0.098 |
| spata18 | 0.274 | nipsnap3a | -0.097 |
| tp73 | 0.272 | fbp1b | -0.097 |
| zgc:109949 | 0.271 | phyhd1 | -0.095 |
| ift27 | 0.271 | lrp2a | -0.095 |
| lrriq1 | 0.271 | aco1 | -0.095 |
| spag6 | 0.270 | slc26a6l | -0.095 |
| si:ch211-81a5.1 | 0.269 | slc16a10 | -0.094 |
| dnaaf4 | 0.268 | smim1 | -0.094 |
| LOC101886896 | 0.267 | rplp2 | -0.094 |
| si:cabz01069015.1 | 0.267 | tktb | -0.094 |