coiled-coil domain containing 106b
ZFIN 
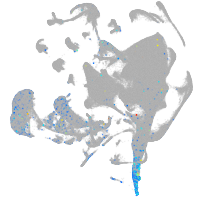
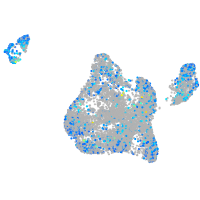
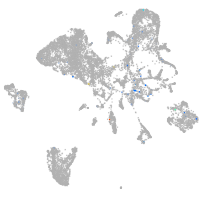
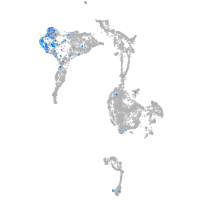
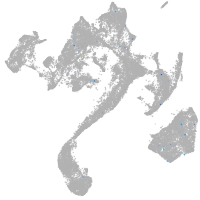
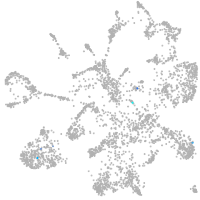
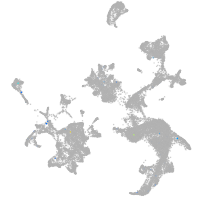
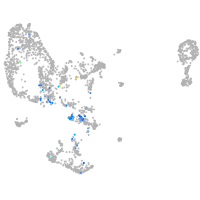
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| flvcr1 | 0.277 | prdx2 | -0.115 |
| cd151l | 0.273 | mif | -0.110 |
| trim109 | 0.271 | si:ch211-139a5.9 | -0.106 |
| LOC101883320 | 0.269 | nme2b.1 | -0.103 |
| BX469925.3 | 0.261 | sod2 | -0.102 |
| chata | 0.250 | prdx6 | -0.102 |
| tgm2a | 0.246 | gapdh | -0.101 |
| si:dkeyp-27c8.2 | 0.237 | gstt1a | -0.099 |
| camta1a | 0.236 | eef1da | -0.098 |
| si:ch211-163l21.7 | 0.233 | mt-nd3 | -0.094 |
| spaca9 | 0.231 | phyhd1 | -0.094 |
| casq1b | 0.229 | COX3 | -0.092 |
| stm | 0.227 | rpl34 | -0.091 |
| cfap52 | 0.223 | cox6a2 | -0.090 |
| BX601648.1 | 0.223 | rplp2 | -0.089 |
| foxj1a | 0.220 | glud1b | -0.088 |
| apeh | 0.219 | dpys | -0.087 |
| LOC101884617 | 0.218 | pvalb1 | -0.087 |
| BX677668.1 | 0.218 | gstk1 | -0.086 |
| daw1 | 0.216 | rps18 | -0.086 |
| alox5b.3 | 0.212 | hoga1 | -0.086 |
| LOC101886811 | 0.211 | tktb | -0.085 |
| lrrc75bb | 0.209 | cat | -0.085 |
| rhof | 0.209 | alpl | -0.084 |
| col8a1a | 0.208 | sod1 | -0.084 |
| si:dkey-33c12.3 | 0.206 | idh1 | -0.083 |
| prxl2b | 0.204 | adh5 | -0.083 |
| id2b | 0.201 | suclg2 | -0.082 |
| GID4 | 0.201 | pnp6 | -0.082 |
| b2m | 0.199 | aldh7a1 | -0.082 |
| ccdc114 | 0.199 | dglucy | -0.082 |
| eno4 | 0.196 | ahcy | -0.082 |
| lrrc74b | 0.195 | si:dkey-16p21.8 | -0.081 |
| tex36 | 0.191 | slc15a2 | -0.080 |
| tctex1d1 | 0.191 | hao1 | -0.080 |