chromobox homolog 1b (HP1 beta homolog Drosophila)
ZFIN 
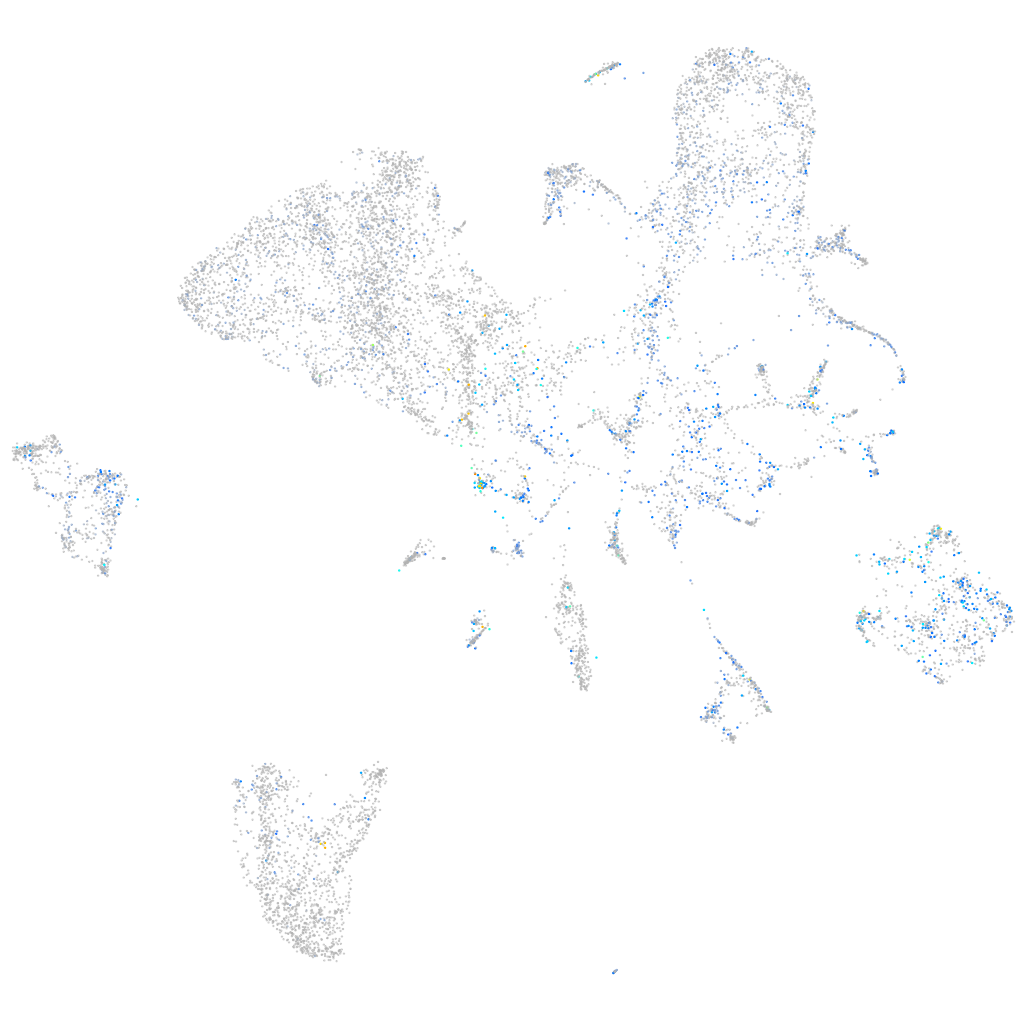
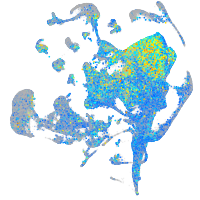
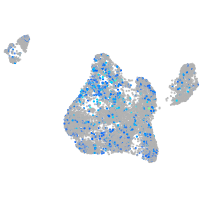
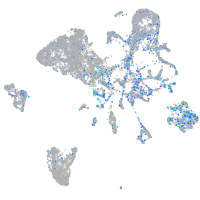
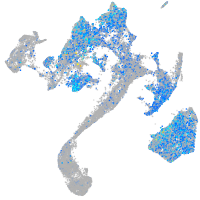
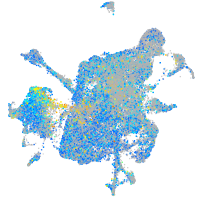
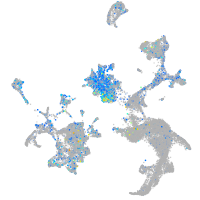
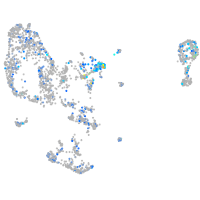
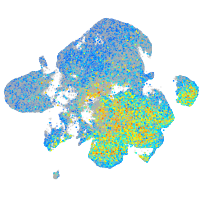
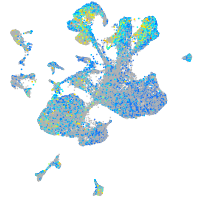
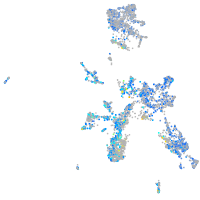
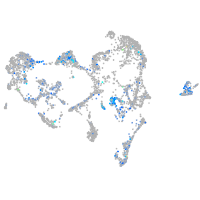
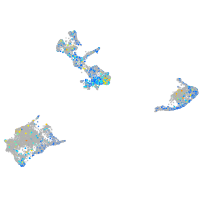
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-222l21.1 | 0.230 | gamt | -0.188 |
| si:ch73-1a9.3 | 0.228 | gatm | -0.179 |
| hmgb1a | 0.226 | gapdh | -0.176 |
| h3f3d | 0.216 | ahcy | -0.169 |
| ptmab | 0.212 | bhmt | -0.161 |
| hmgb3a | 0.211 | mat1a | -0.161 |
| nucks1a | 0.209 | zgc:92744 | -0.159 |
| hnrnpaba | 0.206 | agxtb | -0.152 |
| hmgn6 | 0.206 | pnp4b | -0.152 |
| marcksl1b | 0.200 | dap | -0.151 |
| si:ch211-288g17.3 | 0.199 | aqp12 | -0.150 |
| hmgn7 | 0.199 | fbp1b | -0.148 |
| hmgb1b | 0.199 | gpx4a | -0.147 |
| si:ch73-281n10.2 | 0.198 | nupr1b | -0.146 |
| cirbpa | 0.197 | aldob | -0.143 |
| seta | 0.196 | cx32.3 | -0.142 |
| hmgn2 | 0.196 | aldh6a1 | -0.139 |
| syncrip | 0.196 | apoa4b.1 | -0.139 |
| h2afvb | 0.196 | apoa1b | -0.138 |
| khdrbs1a | 0.196 | grhprb | -0.135 |
| gng3 | 0.194 | abat | -0.135 |
| hnrnpa0b | 0.193 | g6pca.2 | -0.134 |
| hnrnpabb | 0.192 | scp2a | -0.134 |
| hp1bp3 | 0.192 | mgst1.2 | -0.133 |
| cbx3a | 0.192 | ces2 | -0.133 |
| hmgb2b | 0.191 | glud1b | -0.132 |
| hmga1a | 0.190 | aldh7a1 | -0.132 |
| sub1a | 0.187 | afp4 | -0.131 |
| cirbpb | 0.186 | ckba | -0.129 |
| hmgb2a | 0.185 | fabp10a | -0.129 |
| acin1a | 0.185 | ttr | -0.129 |
| chd4a | 0.185 | apoa2 | -0.128 |
| tuba1c | 0.184 | serpina1 | -0.128 |
| anp32a | 0.183 | rbp4 | -0.128 |
| marcksb | 0.182 | etnppl | -0.128 |