"Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase"
ZFIN 
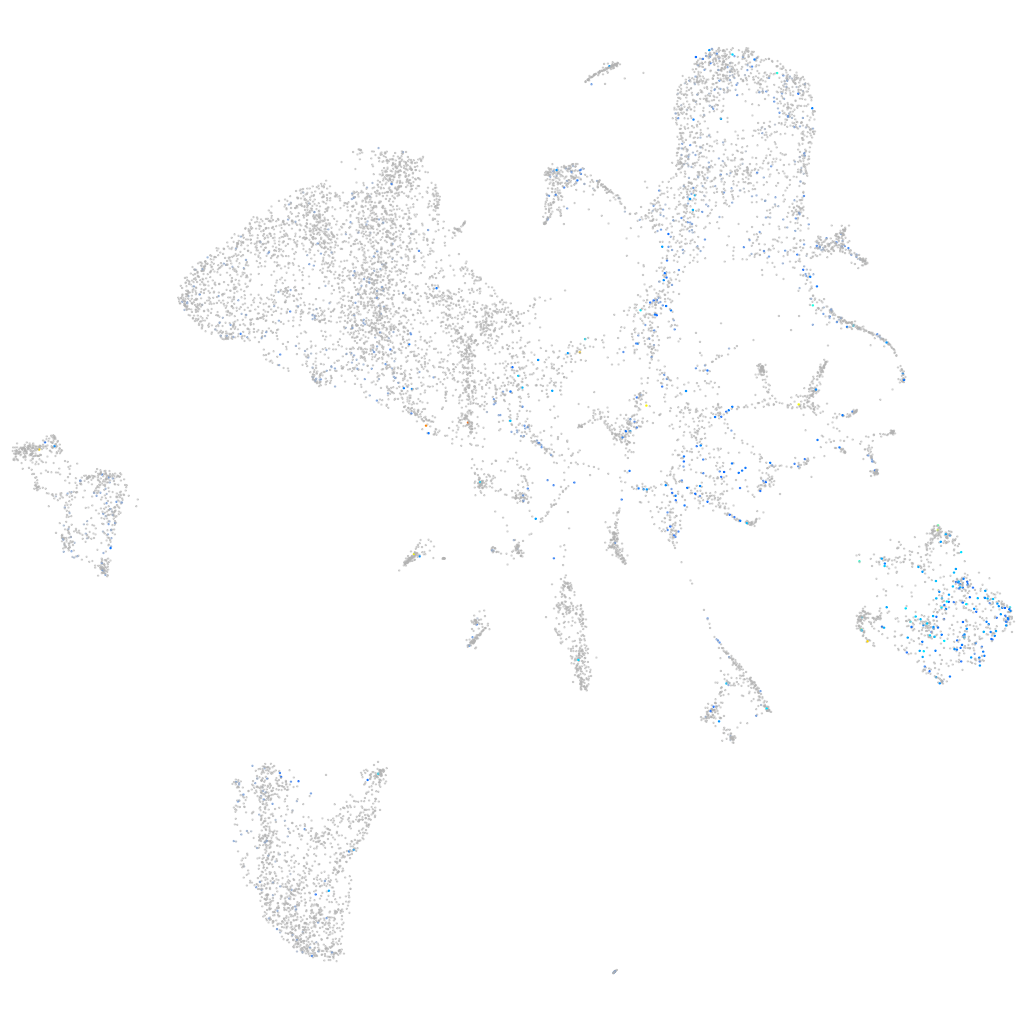
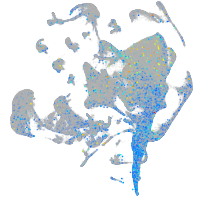
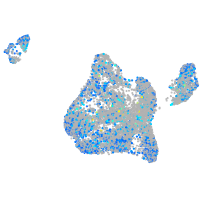
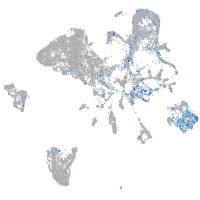
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sox32 | 0.207 | rpl37 | -0.147 |
| hnrnpub | 0.204 | rps10 | -0.137 |
| akap12b | 0.203 | gamt | -0.130 |
| hnrnpa1b | 0.202 | zgc:114188 | -0.126 |
| si:ch211-152c2.3 | 0.199 | bhmt | -0.124 |
| sp5l | 0.195 | rps17 | -0.119 |
| cxcr4a | 0.194 | ahcy | -0.116 |
| nucks1a | 0.194 | zgc:92744 | -0.115 |
| rbm4.3 | 0.192 | gatm | -0.112 |
| dnmt3bb.2 | 0.191 | apoa1b | -0.109 |
| sox17 | 0.189 | agxtb | -0.108 |
| si:dkeyp-2e4.3 | 0.188 | gapdh | -0.107 |
| ilf3b | 0.188 | apoa2 | -0.107 |
| srrm2 | 0.187 | grhprb | -0.107 |
| marcksb | 0.186 | fabp3 | -0.106 |
| LOC100008030 | 0.185 | nme2b.1 | -0.103 |
| vox | 0.185 | mat1a | -0.102 |
| hspb1 | 0.184 | nupr1b | -0.099 |
| top1l | 0.184 | rbp4 | -0.098 |
| nop58 | 0.182 | pnp4b | -0.098 |
| hmga1a | 0.181 | serpina1 | -0.098 |
| syncrip | 0.180 | fabp10a | -0.096 |
| rtf1 | 0.180 | fetub | -0.096 |
| hdgfl2 | 0.180 | hao1 | -0.095 |
| smc1al | 0.180 | eno3 | -0.095 |
| snrpa | 0.180 | sod1 | -0.095 |
| hnrnpabb | 0.179 | ces2 | -0.094 |
| ppig | 0.178 | eef1da | -0.094 |
| seta | 0.178 | aqp12 | -0.094 |
| marcksl1b | 0.178 | agxta | -0.094 |
| sae1 | 0.177 | rbp2b | -0.094 |
| khdrbs1a | 0.177 | suclg1 | -0.093 |
| cx43.4 | 0.177 | pklr | -0.093 |
| hnrnph1l | 0.177 | kng1 | -0.093 |
| NC-002333.4 | 0.177 | apom | -0.093 |