"caspase 8, apoptosis-related cysteine peptidase, like 2 [Source:ZFIN;Acc:ZDB-GENE-070608-2]"
ZFIN 
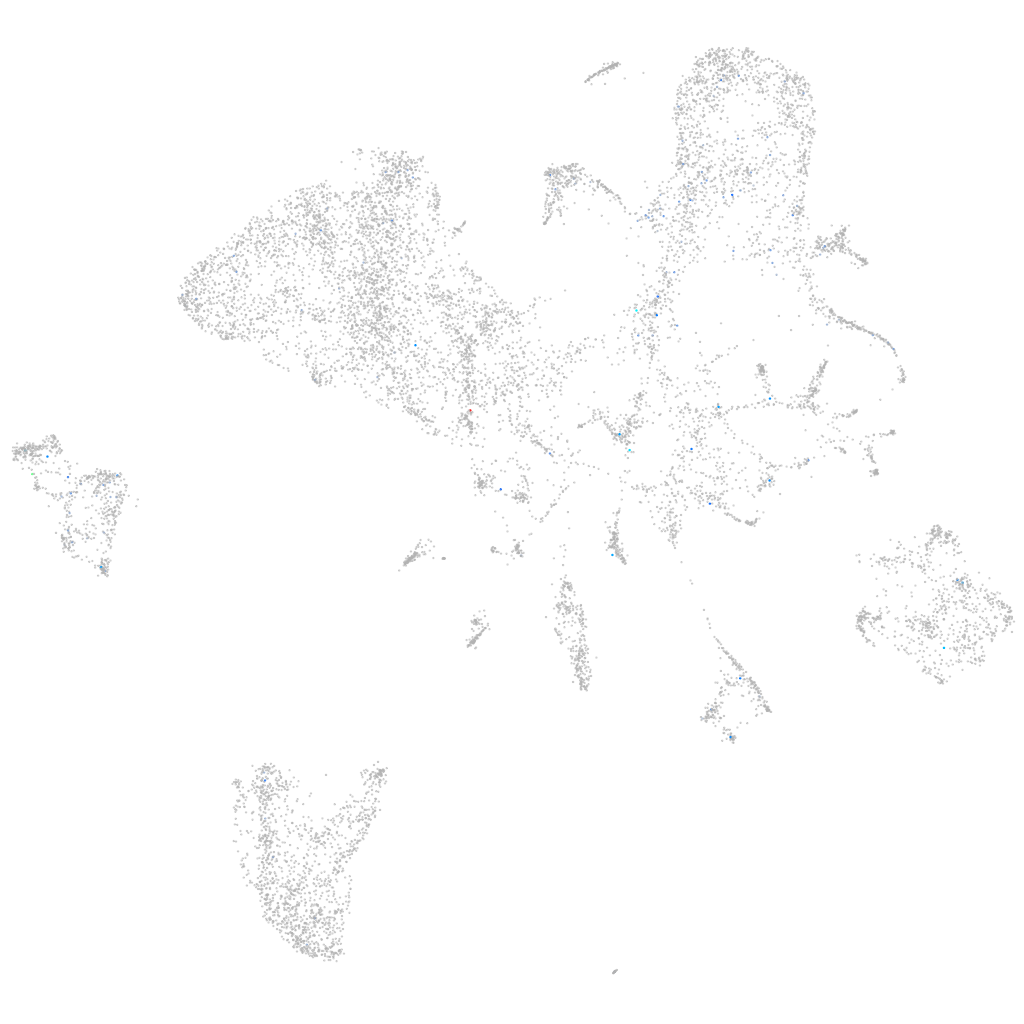
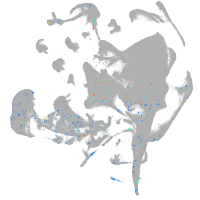
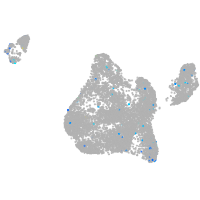
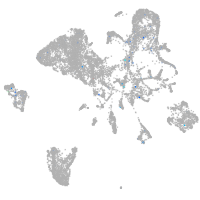
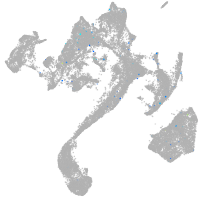
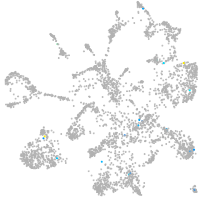
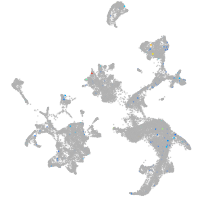
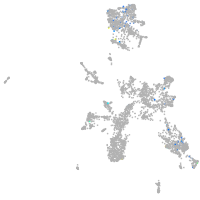
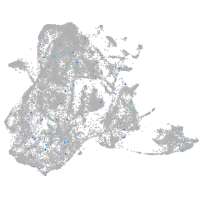
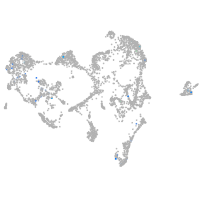
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myo1f | 0.451 | bhmt | -0.047 |
| irf8 | 0.446 | gatm | -0.045 |
| tagapb | 0.381 | aqp12 | -0.041 |
| arhgef3l | 0.359 | gamt | -0.041 |
| igfbp6b | 0.347 | grhprb | -0.037 |
| RF00560 | 0.343 | agxtb | -0.037 |
| ccl34b.1 | 0.321 | gapdh | -0.036 |
| CU984600.2 | 0.296 | zgc:123103 | -0.036 |
| reep6 | 0.224 | serpina1l | -0.035 |
| itgb8 | 0.223 | kng1 | -0.035 |
| gpr31 | 0.222 | mat1a | -0.035 |
| flrt1b | 0.214 | apoa2 | -0.034 |
| BX005183.3 | 0.211 | hao1 | -0.034 |
| wipf1b | 0.204 | serpina1 | -0.034 |
| XLOC-033137 | 0.197 | ttc36 | -0.034 |
| LOC570405.1 | 0.181 | tfa | -0.033 |
| kif21b | 0.180 | hpda | -0.033 |
| LOC100537549 | 0.178 | cfhl4 | -0.033 |
| rps6ka3b | 0.160 | ces2 | -0.033 |
| XLOC-042595 | 0.159 | agxta | -0.033 |
| herc3 | 0.155 | ambp | -0.033 |
| zbtb7b | 0.152 | vtnb | -0.033 |
| CU914776.1 | 0.149 | fgg | -0.032 |
| BX572627.1 | 0.146 | fgb | -0.032 |
| CR376738.1 | 0.144 | rbp4 | -0.032 |
| ogfr | 0.144 | afp4 | -0.032 |
| bace1 | 0.141 | g6pca.2 | -0.032 |
| znf1038 | 0.139 | LOC110437731 | -0.032 |
| CABZ01080371.1 | 0.138 | c8g | -0.031 |
| bzw2 | 0.136 | si:ch211-270n8.1 | -0.031 |
| si:dkey-9l20.3 | 0.133 | ttr | -0.031 |
| pcp4l1 | 0.130 | pygl | -0.031 |
| asphd2 | 0.129 | gc | -0.031 |
| lmo3 | 0.127 | c9 | -0.030 |
| si:ch73-27e22.1 | 0.124 | f2 | -0.030 |