casc3 exon junction complex subunit
ZFIN 
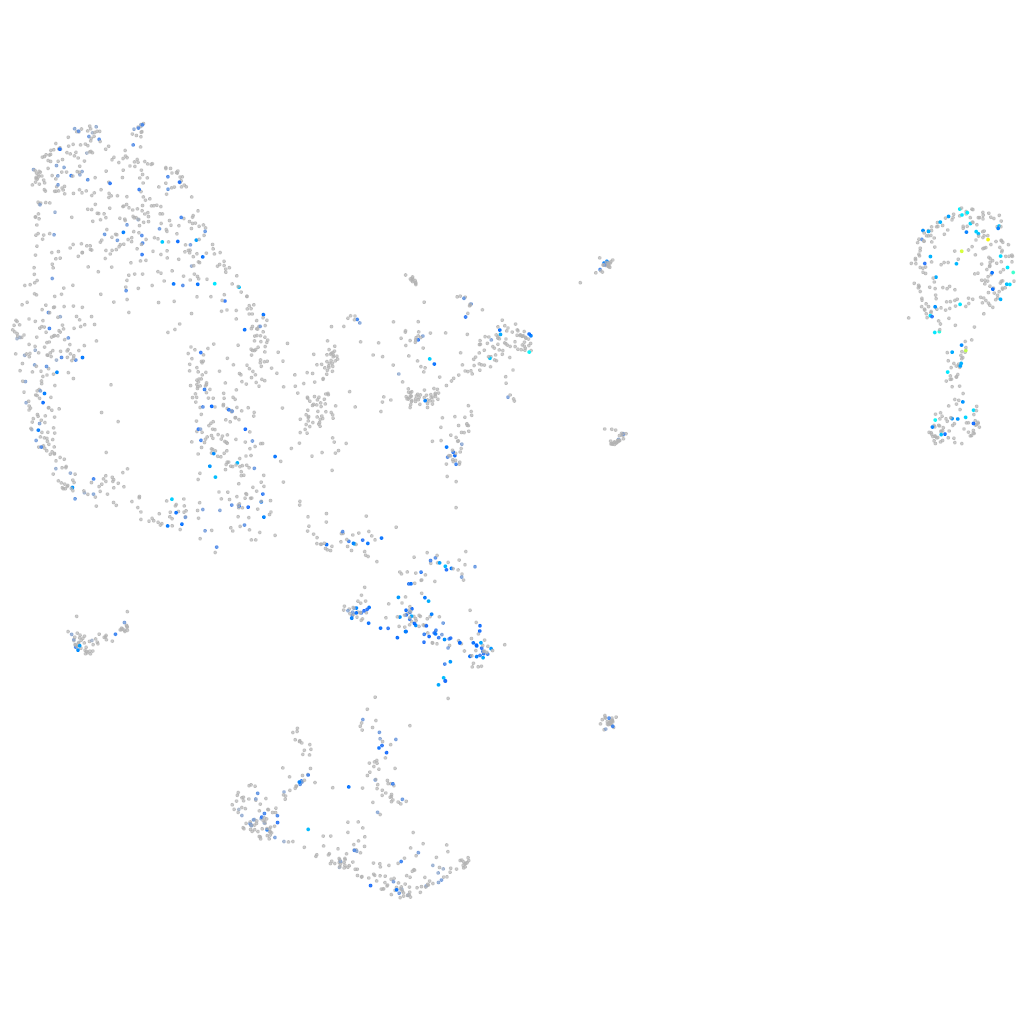
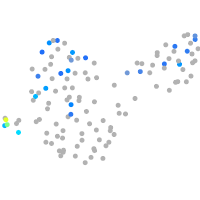
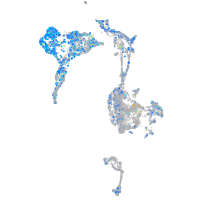
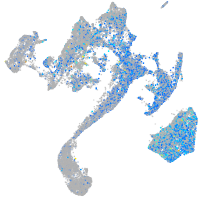
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.221 | si:ch211-139a5.9 | -0.189 |
| BX927258.1 | 0.214 | eno3 | -0.171 |
| hmga1a | 0.211 | prdx2 | -0.157 |
| nucks1a | 0.210 | gstp1 | -0.154 |
| XLOC-039121 | 0.208 | aldob | -0.153 |
| hnrnpabb | 0.208 | vdac3 | -0.148 |
| ddx39ab | 0.205 | gapdh | -0.143 |
| cxcr4b | 0.203 | si:dkey-16p21.8 | -0.142 |
| setb | 0.203 | grhprb | -0.137 |
| cbx3a | 0.201 | gstr | -0.137 |
| h2afvb | 0.201 | agxta | -0.136 |
| ptges3b | 0.197 | aldh6a1 | -0.136 |
| si:ch211-222l21.1 | 0.195 | gstt1a | -0.135 |
| crtap | 0.195 | sod1 | -0.134 |
| lbx2 | 0.194 | ldhba | -0.134 |
| si:ch211-288g17.3 | 0.193 | nit2 | -0.131 |
| hnrnph1l | 0.192 | ahcy | -0.131 |
| rbm4.3 | 0.192 | rpl37 | -0.130 |
| smarca4a | 0.191 | pvalb1 | -0.129 |
| seta | 0.191 | glud1b | -0.129 |
| ilf3b | 0.190 | sod2 | -0.127 |
| si:ch73-281n10.2 | 0.189 | pvalb2 | -0.127 |
| snrpb | 0.188 | hoga1 | -0.126 |
| mcm2 | 0.188 | gamt | -0.126 |
| hnrnpaba | 0.185 | pklr | -0.125 |
| apoeb | 0.185 | elovl1b | -0.125 |
| dnaaf4 | 0.183 | cox7a1 | -0.125 |
| snrpd2 | 0.183 | eef1da | -0.125 |
| XLOC-032526 | 0.182 | atp5mc3b | -0.124 |
| hmgb2a | 0.181 | cox6b1 | -0.124 |
| sap18 | 0.181 | atp5l | -0.124 |
| khdrbs1a | 0.180 | atp5f1e | -0.123 |
| myef2 | 0.180 | fbp1b | -0.122 |
| wu:fb97g03 | 0.180 | cox6c | -0.121 |
| hmgn6 | 0.180 | suclg1 | -0.121 |