"calmodulin regulated spectrin-associated protein family, member 3"
ZFIN 
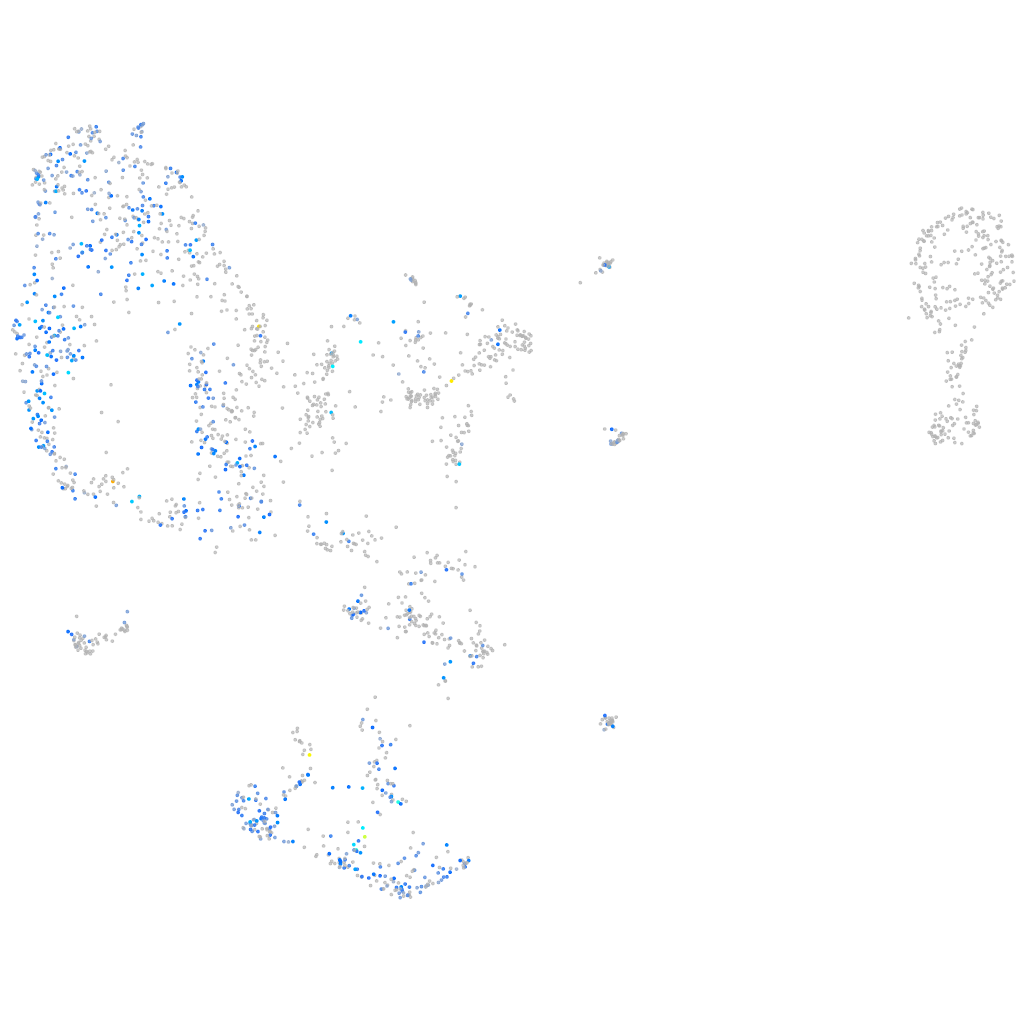
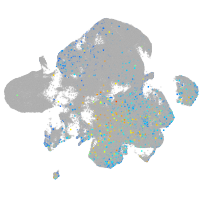
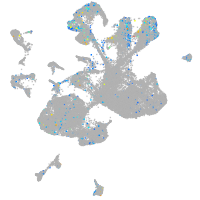
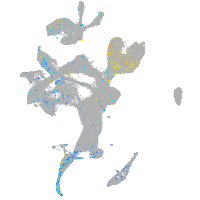
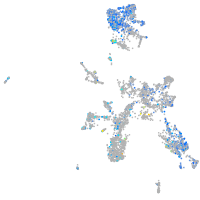
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh17 | 0.336 | marcksl1b | -0.247 |
| chp1 | 0.313 | marcksb | -0.235 |
| mt-nd4 | 0.292 | wu:fb97g03 | -0.227 |
| mt-nd2 | 0.287 | hspb1 | -0.222 |
| atp1b1a | 0.285 | hmgb2b | -0.216 |
| si:ch211-139a5.9 | 0.284 | hnrnpaba | -0.216 |
| aldh1l1 | 0.284 | ptmab | -0.213 |
| glud1b | 0.282 | si:ch73-1a9.3 | -0.211 |
| sgk2a | 0.279 | apoeb | -0.208 |
| sgk1 | 0.276 | hmgn6 | -0.208 |
| mt-co1 | 0.276 | hnrnpabb | -0.207 |
| mt-atp6 | 0.275 | hmgb1b | -0.205 |
| exoc3l2b | 0.273 | si:ch211-222l21.1 | -0.204 |
| lrrc66 | 0.271 | h3f3d | -0.198 |
| mt-cyb | 0.269 | nucks1a | -0.197 |
| mt-nd1 | 0.269 | apoc1 | -0.195 |
| atp5fa1 | 0.269 | acin1a | -0.194 |
| igfbp5b | 0.267 | seta | -0.192 |
| si:dkey-16p21.8 | 0.267 | si:ch73-281n10.2 | -0.191 |
| ldhba | 0.265 | h2afvb | -0.190 |
| suclg1 | 0.264 | hmgn7 | -0.187 |
| eno3 | 0.264 | hnrnpa1a | -0.187 |
| aldh6a1 | 0.264 | hmga1a | -0.184 |
| cdaa | 0.263 | cx43.4 | -0.184 |
| atp5f1b | 0.263 | syncrip | -0.184 |
| COX3 | 0.262 | hmgn2 | -0.184 |
| slc13a3 | 0.261 | nop58 | -0.182 |
| cox6a2 | 0.260 | snrpd2 | -0.181 |
| mdh1aa | 0.260 | ptges3b | -0.181 |
| slc25a5 | 0.259 | fbl | -0.181 |
| atp1a1a.4 | 0.258 | akap12b | -0.179 |
| kcnj13 | 0.258 | anp32b | -0.178 |
| rbm47 | 0.256 | sumo3b | -0.174 |
| etnk1 | 0.253 | cbx3a | -0.174 |
| mt-co2 | 0.252 | cirbpa | -0.174 |