calcium/calmodulin-dependent protein kinase 1Da
ZFIN 
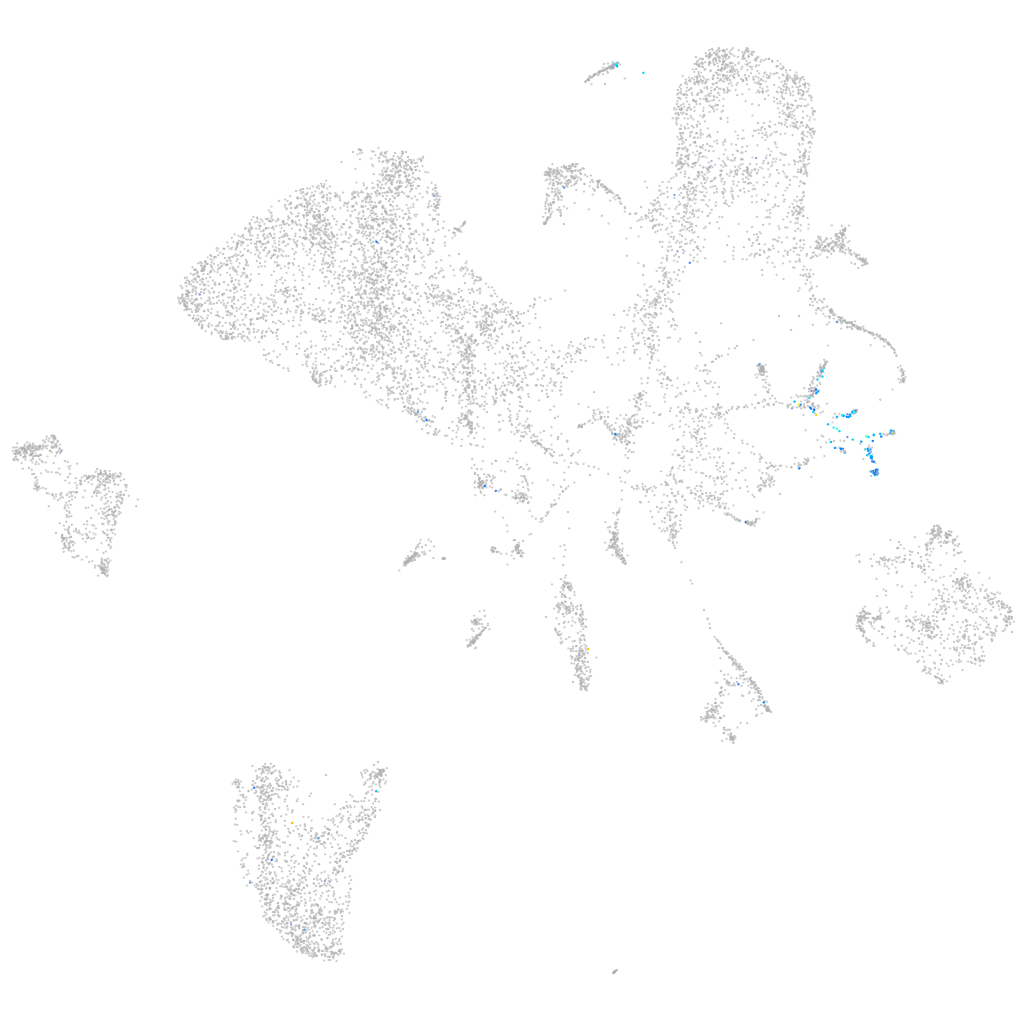
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.467 | aldob | -0.109 |
| si:dkey-153k10.9 | 0.431 | ahcy | -0.108 |
| rprmb | 0.413 | gapdh | -0.108 |
| pcsk1nl | 0.411 | gstp1 | -0.096 |
| SLC5A10 | 0.410 | rps20 | -0.095 |
| scg3 | 0.401 | gstr | -0.094 |
| pax6b | 0.400 | gamt | -0.094 |
| kcnj11 | 0.399 | aldh6a1 | -0.093 |
| isl1 | 0.398 | eno3 | -0.092 |
| neurod1 | 0.391 | aldh9a1a.1 | -0.089 |
| dkk3b | 0.389 | gstt1a | -0.084 |
| scg2a | 0.388 | fbp1b | -0.081 |
| ndufa4l2a | 0.380 | si:dkey-16p21.8 | -0.081 |
| pcsk1 | 0.377 | apoa4b.1 | -0.081 |
| rims2a | 0.376 | nupr1b | -0.079 |
| pcsk2 | 0.372 | apoa1b | -0.079 |
| cntnap2a | 0.371 | eef1da | -0.079 |
| tspan7b | 0.358 | ppa1b | -0.078 |
| cacna1c | 0.355 | krt18a.1 | -0.078 |
| syt1a | 0.341 | apoc2 | -0.078 |
| kcnk9 | 0.341 | gcshb | -0.077 |
| mir7a-1 | 0.335 | cebpd | -0.077 |
| ptn | 0.332 | gatm | -0.077 |
| si:ch73-287m6.1 | 0.332 | scp2a | -0.077 |
| insm1a | 0.327 | hspe1 | -0.076 |
| gcgb | 0.325 | fabp3 | -0.076 |
| scg2b | 0.320 | mat1a | -0.075 |
| vat1 | 0.320 | cx32.3 | -0.075 |
| adm2a | 0.318 | glud1b | -0.075 |
| CR774179.5 | 0.314 | bhmt | -0.075 |
| LOC571123 | 0.313 | agxta | -0.075 |
| jph3 | 0.312 | hdlbpa | -0.075 |
| ins | 0.310 | afp4 | -0.074 |
| slc30a2 | 0.309 | acadm | -0.073 |
| elfn1a | 0.308 | eif4ebp1 | -0.073 |