calcium homeostasis modulator family member 6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 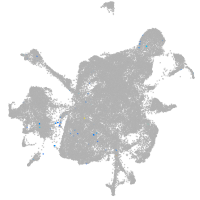 |
hematopoietic / vasculature 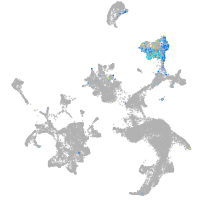 |
pronephros 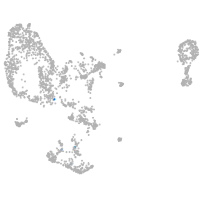 |
neural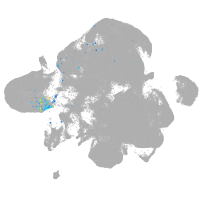 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fermt1 | 0.170 | rpl37 | -0.066 |
| tagln2 | 0.162 | rps10 | -0.064 |
| mir205 | 0.160 | zgc:114188 | -0.062 |
| epcam | 0.138 | ptmaa | -0.062 |
| cdh1 | 0.129 | h3f3a | -0.058 |
| tp63 | 0.129 | hnrnpa0l | -0.054 |
| dlx3b | 0.128 | tuba1c | -0.053 |
| myh9a | 0.128 | fabp3 | -0.053 |
| cldn7b | 0.122 | nova2 | -0.052 |
| f11r.1 | 0.113 | marcksl1a | -0.048 |
| hpgd | 0.111 | rps17 | -0.046 |
| arhgef5 | 0.110 | elavl3 | -0.043 |
| si:ch211-199g17.2 | 0.109 | gpm6aa | -0.043 |
| nkx2.7 | 0.104 | rtn1a | -0.043 |
| grhl2b | 0.098 | si:ch1073-429i10.3.1 | -0.042 |
| apoc1 | 0.096 | CR383676.1 | -0.040 |
| apoeb | 0.095 | stmn1b | -0.039 |
| tnfaip8l2b | 0.092 | zgc:158463 | -0.039 |
| F2RL3 | 0.088 | tmsb4x | -0.039 |
| pak2a | 0.088 | ppiab | -0.038 |
| LECT2 | 0.086 | ckbb | -0.037 |
| pdlim1 | 0.085 | nme2b.1 | -0.036 |
| cd9b | 0.083 | mdkb | -0.035 |
| CU469568.1 | 0.082 | cct2 | -0.034 |
| zgc:171775 | 0.082 | celf2 | -0.034 |
| cavin2b | 0.082 | tubb5 | -0.034 |
| nid2a | 0.081 | tmsb | -0.033 |
| krt8 | 0.080 | h3f3c | -0.033 |
| CT030188.1 | 0.080 | tuba1a | -0.033 |
| foxi1 | 0.080 | zc4h2 | -0.032 |
| cldni | 0.080 | gng3 | -0.032 |
| cnn2 | 0.080 | rnasekb | -0.032 |
| itga3b | 0.080 | calm1a | -0.032 |
| capn8 | 0.079 | gpm6ab | -0.031 |
| dspa | 0.079 | gapdhs | -0.031 |