"calcitonin/calcitonin-related polypeptide, alpha"
ZFIN 
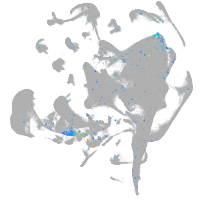
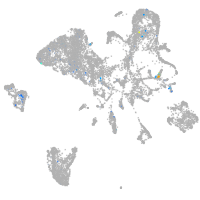
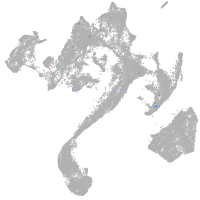
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lgi2a | 0.588 | si:ch73-1a9.3 | -0.030 |
| slc22a6l | 0.582 | ap1s1 | -0.028 |
| LOC101883560 | 0.442 | sem1 | -0.026 |
| CR792422.2 | 0.386 | ptmab | -0.026 |
| LOC101885328 | 0.330 | btf3l4 | -0.026 |
| si:ch73-236c18.5 | 0.330 | cct7 | -0.025 |
| CR394546.2 | 0.330 | p4hb | -0.025 |
| CU929037.3 | 0.330 | tmem11 | -0.025 |
| LOC110439567 | 0.330 | psma6a | -0.024 |
| LOC101882039 | 0.319 | vps28 | -0.024 |
| si:ch211-202n12.2 | 0.292 | si:dkey-151g10.6 | -0.024 |
| pacrg | 0.285 | tmed4 | -0.024 |
| CR936240.1 | 0.276 | nsfl1c | -0.023 |
| LOC100334356 | 0.276 | canx | -0.023 |
| LOC108183454 | 0.275 | phf5a | -0.023 |
| BX530017.2 | 0.263 | tmem258 | -0.023 |
| BX119915.2 | 0.249 | psma5 | -0.023 |
| si:ch211-214k5.6 | 0.240 | sap18 | -0.023 |
| fhl5 | 0.236 | psma3 | -0.022 |
| drgx | 0.232 | taf3 | -0.022 |
| CU984600.2 | 0.222 | si:dkey-46a10.3 | -0.022 |
| XLOC-014033 | 0.213 | rps21 | -0.022 |
| trim35-3 | 0.210 | sart1 | -0.021 |
| CR846087.3 | 0.208 | bzw1a | -0.021 |
| il13 | 0.203 | sept8a | -0.021 |
| sugct | 0.203 | hdac1 | -0.021 |
| cldnd | 0.201 | pim3 | -0.021 |
| LOC101884668 | 0.193 | ccdc25 | -0.021 |
| npy2r | 0.192 | nav2b | -0.021 |
| scml4 | 0.190 | emc7 | -0.021 |
| trpa1b | 0.182 | prkcsh | -0.021 |
| FP102806.1 | 0.177 | gcsha | -0.021 |
| LOC110437714 | 0.172 | fam133b | -0.020 |
| tcap | 0.171 | hmgb1a | -0.020 |
| slmapb | 0.170 | hgs | -0.020 |