Ca2+-dependent activator protein for secretion b
ZFIN 
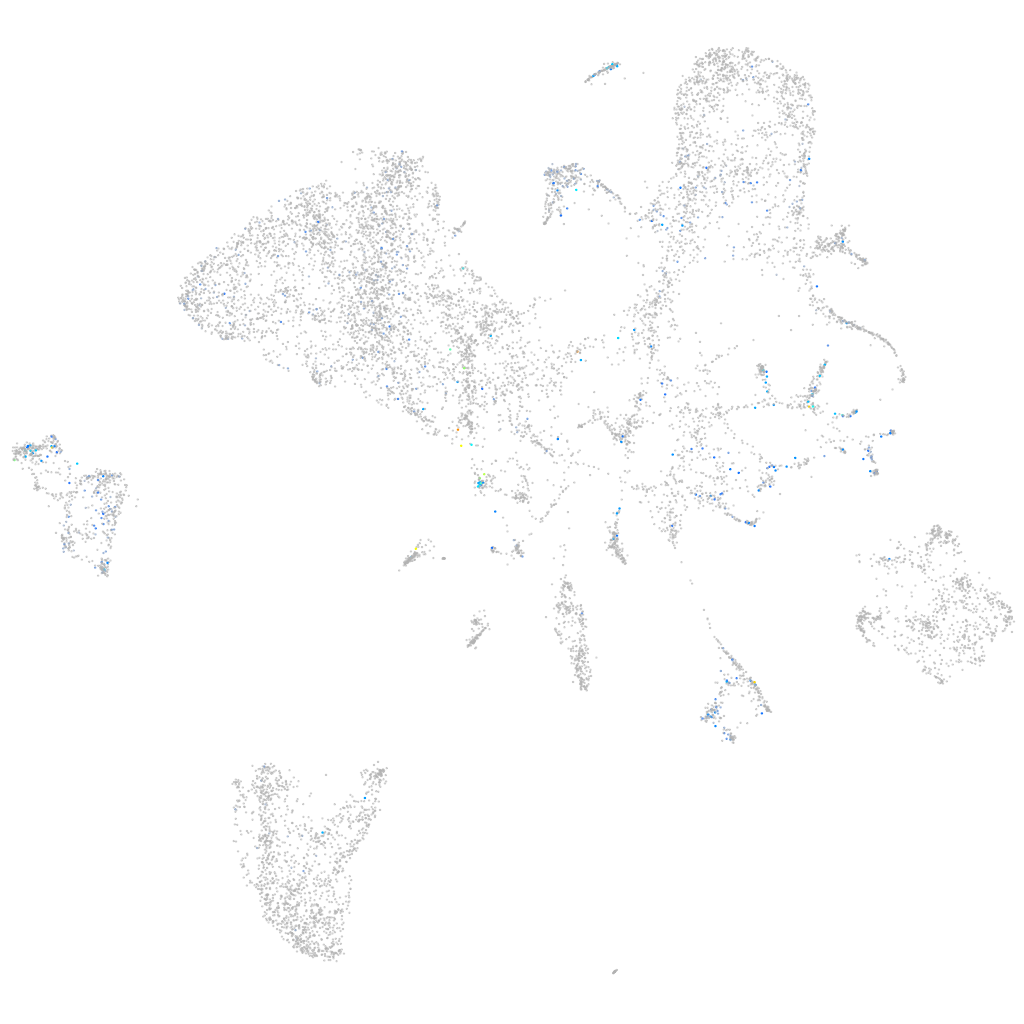
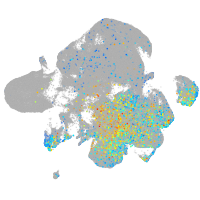
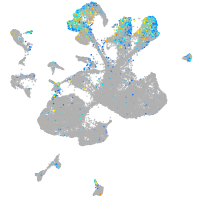
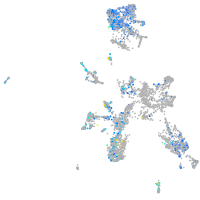
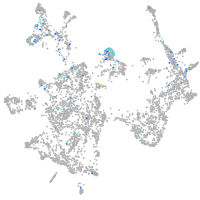
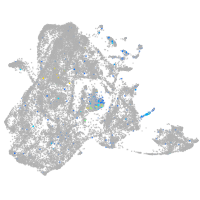
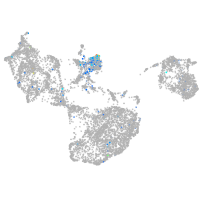
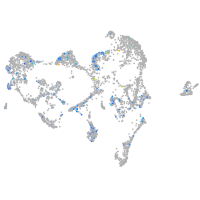
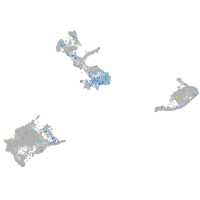
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mtus2a | 0.191 | gatm | -0.096 |
| slitrk5a | 0.162 | gapdh | -0.088 |
| syt1a | 0.158 | gamt | -0.086 |
| myt1la | 0.155 | ahcy | -0.084 |
| snap25b | 0.151 | bhmt | -0.072 |
| calm1b | 0.149 | aqp12 | -0.072 |
| gapdhs | 0.148 | mat1a | -0.071 |
| si:dkey-210j14.3 | 0.148 | cx32.3 | -0.066 |
| elavl4 | 0.148 | apoa4b.1 | -0.065 |
| tbkbp1 | 0.146 | fbp1b | -0.065 |
| crygm2d3 | 0.139 | nupr1b | -0.062 |
| camk2n1a | 0.138 | slc16a10 | -0.062 |
| id4 | 0.136 | dap | -0.061 |
| rs1b | 0.136 | afp4 | -0.060 |
| sncb | 0.135 | gstr | -0.060 |
| urgcp | 0.135 | ppdpfa | -0.058 |
| stx1b | 0.134 | glud1b | -0.058 |
| pcdh1a | 0.133 | gcshb | -0.058 |
| rgs20 | 0.128 | apoc1 | -0.057 |
| mir199-2 | 0.127 | cdo1 | -0.057 |
| LOC100149700 | 0.126 | scp2a | -0.057 |
| gpm6aa | 0.126 | apoc2 | -0.056 |
| kcna1a | 0.126 | tram1 | -0.055 |
| necap1 | 0.126 | eif4ebp3 | -0.055 |
| atp1a3a | 0.125 | rgrb | -0.055 |
| gnb1a | 0.125 | dhrs9 | -0.055 |
| calm1a | 0.124 | adka | -0.055 |
| stxbp1a | 0.124 | hspe1 | -0.054 |
| kdm6bb | 0.124 | apobb.1 | -0.054 |
| fev | 0.123 | zgc:92744 | -0.054 |
| slc6a1a | 0.123 | etnppl | -0.054 |
| ppp1r3aa | 0.122 | pnp4b | -0.054 |
| drd2a | 0.121 | aldh6a1 | -0.053 |
| scg2a | 0.121 | gstt1a | -0.053 |
| sh3bp2 | 0.121 | sec61b | -0.052 |