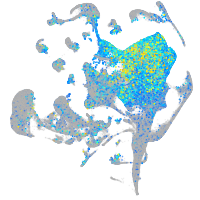
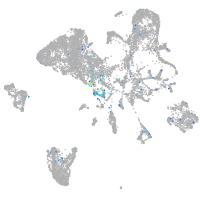
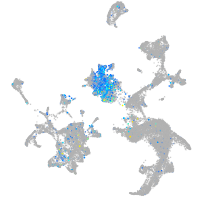
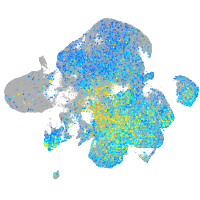
cell adhesion molecule 3
ZFIN 
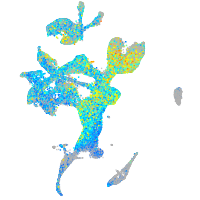
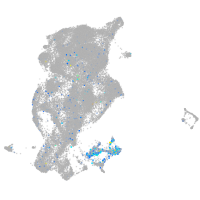
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.487 | tspan36 | -0.177 |
| nova2 | 0.417 | gpr143 | -0.172 |
| rnasekb | 0.415 | syngr1a | -0.169 |
| epb41a | 0.390 | smim29 | -0.157 |
| sypb | 0.388 | paics | -0.154 |
| tuba1c | 0.375 | gstp1 | -0.151 |
| rtn1a | 0.365 | rab32a | -0.151 |
| ptmaa | 0.358 | cst14a.2 | -0.149 |
| foxg1b | 0.357 | si:dkey-21a6.5 | -0.147 |
| snap25b | 0.351 | akr1b1 | -0.137 |
| hmgb1b | 0.347 | actb2 | -0.136 |
| atp6v0cb | 0.341 | impdh1b | -0.136 |
| rorb | 0.338 | pts | -0.134 |
| stmn1b | 0.336 | gmps | -0.134 |
| si:ch73-1a9.3 | 0.336 | pfn1 | -0.134 |
| neurod4 | 0.331 | pttg1ipb | -0.133 |
| tmeff1b | 0.330 | agtrap | -0.133 |
| vamp2 | 0.327 | glulb | -0.131 |
| ptmab | 0.325 | slc2a15a | -0.131 |
| atp1a3a | 0.324 | pcbd1 | -0.130 |
| cspg5a | 0.322 | rnaseka | -0.129 |
| gng3 | 0.321 | qdpra | -0.128 |
| tox | 0.320 | eno3 | -0.127 |
| crx | 0.317 | ahnak | -0.125 |
| sncb | 0.315 | zgc:110239 | -0.125 |
| gpm6ba | 0.314 | LOC103910009 | -0.124 |
| elavl3 | 0.311 | slc45a2 | -0.123 |
| nsg2 | 0.310 | bace2 | -0.122 |
| scrt2 | 0.309 | prps1a | -0.122 |
| ndrg4 | 0.309 | trpm1b | -0.122 |
| marcksl1a | 0.309 | rab38 | -0.121 |
| si:ch211-222l21.1 | 0.307 | zgc:110339 | -0.120 |
| syt1a | 0.304 | sdc4 | -0.120 |
| olfm1b | 0.302 | rabl6b | -0.119 |
| elavl4 | 0.301 | prdx6 | -0.119 |