"calcium channel, voltage-dependent, alpha 2/delta subunit 2a"
ZFIN 
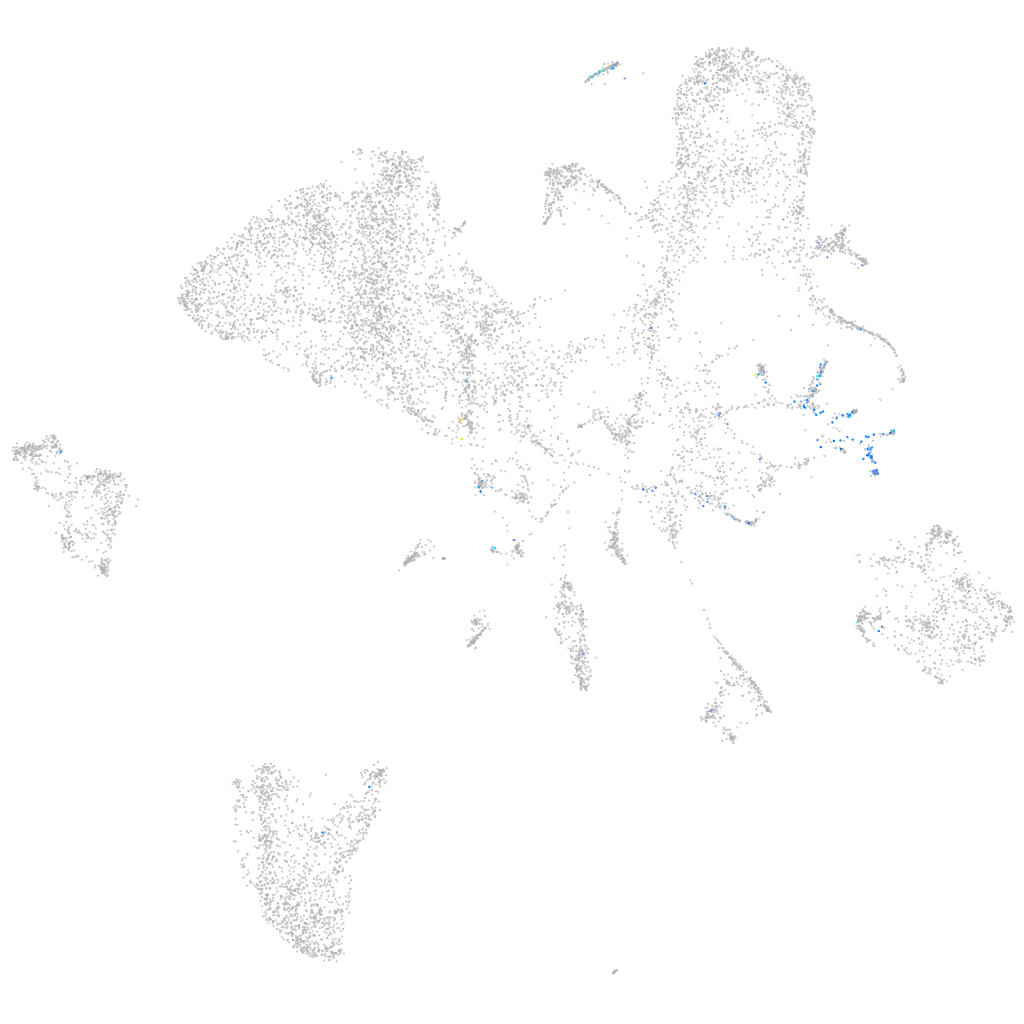
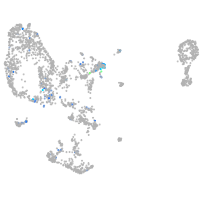
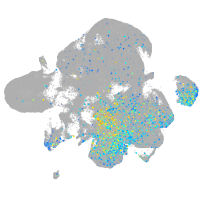
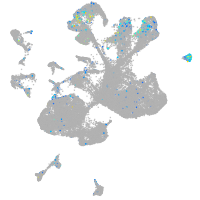
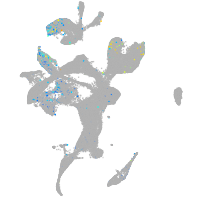
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.320 | ahcy | -0.111 |
| cpe | 0.315 | gapdh | -0.105 |
| scgn | 0.314 | gamt | -0.100 |
| rims2a | 0.310 | gatm | -0.088 |
| neurod1 | 0.300 | fbp1b | -0.085 |
| syt1a | 0.288 | aldh6a1 | -0.084 |
| pcsk1 | 0.279 | rps20 | -0.082 |
| isl1 | 0.276 | mat1a | -0.082 |
| pcsk2 | 0.272 | aldh7a1 | -0.082 |
| SLC5A10 | 0.270 | gstp1 | -0.082 |
| kcnj11 | 0.266 | fabp3 | -0.082 |
| rprmb | 0.265 | gnmt | -0.080 |
| pax6b | 0.262 | gstt1a | -0.079 |
| scg2a | 0.261 | aldob | -0.078 |
| pcsk1nl | 0.256 | apoa4b.1 | -0.078 |
| tspan7b | 0.247 | nupr1b | -0.077 |
| gpc1a | 0.245 | cx32.3 | -0.076 |
| vat1 | 0.243 | eef1da | -0.076 |
| si:dkey-153k10.9 | 0.242 | bhmt | -0.075 |
| sst1.1 | 0.241 | hdlbpa | -0.075 |
| dkk3b | 0.237 | scp2a | -0.075 |
| id4 | 0.233 | eno3 | -0.074 |
| tmem145 | 0.231 | ckba | -0.073 |
| insm1a | 0.231 | cebpd | -0.072 |
| cntnap2a | 0.230 | shmt1 | -0.072 |
| camk1da | 0.229 | gstr | -0.072 |
| CABZ01072563.1 | 0.226 | agxtb | -0.072 |
| kcnk9 | 0.226 | mgst1.2 | -0.072 |
| vamp2 | 0.226 | cox7a1 | -0.072 |
| gria2b | 0.225 | apoc2 | -0.071 |
| mir7a-1 | 0.225 | afp4 | -0.071 |
| si:ch211-264f5.8 | 0.224 | g6pca.2 | -0.071 |
| rnasekb | 0.224 | aqp12 | -0.070 |
| aldocb | 0.222 | apoa1b | -0.070 |
| syt11a | 0.220 | agxta | -0.070 |