"calcium channel, voltage-dependent, L type, alpha 1C subunit"
ZFIN 
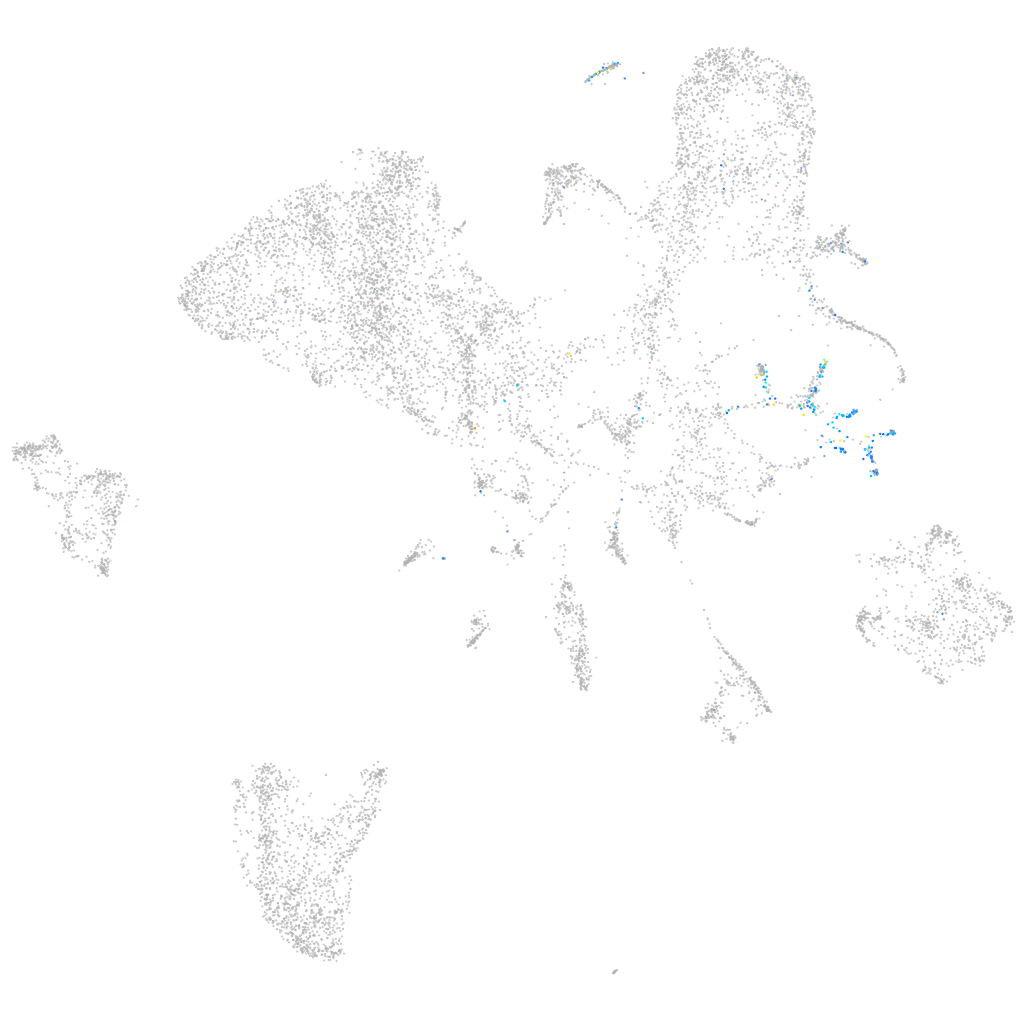
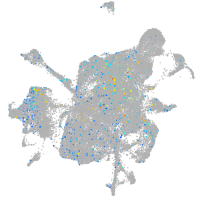
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.484 | ahcy | -0.132 |
| neurod1 | 0.447 | gapdh | -0.125 |
| pax6b | 0.431 | gamt | -0.122 |
| mir7a-1 | 0.427 | hdlbpa | -0.114 |
| scgn | 0.427 | gatm | -0.110 |
| si:dkey-153k10.9 | 0.413 | aldob | -0.109 |
| syt1a | 0.395 | bhmt | -0.108 |
| kcnj11 | 0.394 | fabp3 | -0.107 |
| tspan7b | 0.391 | nupr1b | -0.103 |
| pcsk1nl | 0.385 | mat1a | -0.102 |
| pcsk1 | 0.381 | si:dkey-16p21.8 | -0.102 |
| isl1 | 0.372 | aldh6a1 | -0.101 |
| rims2a | 0.368 | glud1b | -0.100 |
| rprmb | 0.364 | gstr | -0.098 |
| insm1a | 0.359 | aqp12 | -0.098 |
| camk1da | 0.355 | apoa1b | -0.096 |
| scg2a | 0.351 | apoc1 | -0.096 |
| c2cd4a | 0.342 | apoa4b.1 | -0.095 |
| pcsk2 | 0.336 | fbp1b | -0.095 |
| kcnk9 | 0.333 | cx32.3 | -0.095 |
| mir375-2 | 0.333 | aldh7a1 | -0.094 |
| vat1 | 0.332 | agxtb | -0.093 |
| arxa | 0.326 | eif4ebp1 | -0.092 |
| gpc1a | 0.325 | agxta | -0.092 |
| rasd4 | 0.322 | rps20 | -0.091 |
| dkk3b | 0.322 | gstp1 | -0.091 |
| nkx2.2a | 0.317 | apoc2 | -0.091 |
| id4 | 0.312 | gnmt | -0.090 |
| gcgb | 0.311 | afp4 | -0.089 |
| fam49bb | 0.310 | gstt1a | -0.089 |
| nucb2b | 0.306 | adka | -0.088 |
| vamp2 | 0.306 | eno3 | -0.088 |
| insm1b | 0.306 | tktb | -0.087 |
| pygma | 0.305 | apoa2 | -0.086 |
| tmem63c | 0.302 | mgst1.2 | -0.086 |