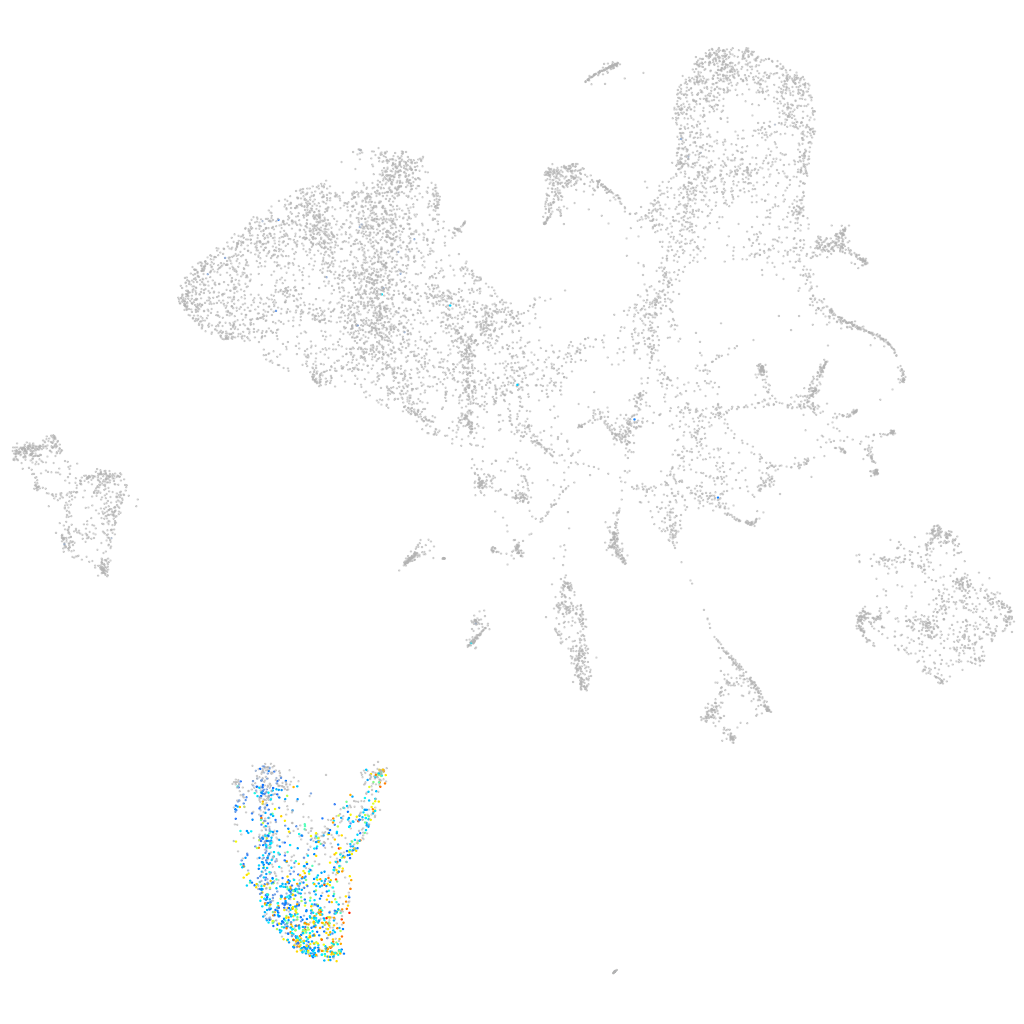
six-cysteine containing astacin protease 3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| c6ast4 | 0.745 | slc25a5 | -0.408 |
| zgc:136461 | 0.722 | rps23 | -0.404 |
| ela2 | 0.722 | prdx2 | -0.376 |
| cpa1 | 0.721 | btf3 | -0.376 |
| amy2a | 0.720 | naca | -0.352 |
| ela2l | 0.718 | rps9 | -0.339 |
| sycn.2 | 0.718 | hsp90ab1 | -0.328 |
| ela3l | 0.716 | rps15a | -0.322 |
| cel.2 | 0.715 | actb2 | -0.321 |
| zgc:92137 | 0.715 | cox6b2 | -0.319 |
| zgc:112160 | 0.711 | rpl19 | -0.311 |
| cpa5 | 0.711 | rpl7 | -0.311 |
| prss59.1 | 0.710 | rps5 | -0.311 |
| cpa4 | 0.708 | rps7 | -0.309 |
| CELA1 (1 of many) | 0.708 | rpl7a | -0.300 |
| cel.1 | 0.708 | fkbp1aa | -0.299 |
| ctrb1 | 0.706 | ubb | -0.296 |
| cpb1 | 0.706 | rpl8 | -0.293 |
| ctrl | 0.697 | rps24 | -0.293 |
| prss1 | 0.695 | mgst3b | -0.291 |
| prss59.2 | 0.691 | fthl27 | -0.291 |
| si:ch211-240l19.5 | 0.689 | rpl13a | -0.290 |
| si:dkey-14d8.6 | 0.680 | calm2b | -0.289 |
| erp27 | 0.680 | rpl12 | -0.288 |
| apoda.2 | 0.679 | gstp1 | -0.287 |
| fep15 | 0.675 | pgrmc1 | -0.281 |
| endou | 0.672 | rpl28 | -0.278 |
| pdia2 | 0.669 | rpl26 | -0.276 |
| spink4 | 0.663 | tpi1b | -0.273 |
| pla2g1b | 0.661 | slc25a3b | -0.271 |
| si:ch211-240l19.6 | 0.656 | rps4x | -0.271 |
| si:ch73-44m9.5 | 0.619 | rpl10a | -0.270 |
| si:dkey-266f7.9 | 0.616 | eif3f | -0.270 |
| si:dkey-14d8.7 | 0.603 | aldob | -0.270 |
| tmed6 | 0.572 | rps3a | -0.270 |