C2 calcium dependent domain containing 4A
ZFIN 
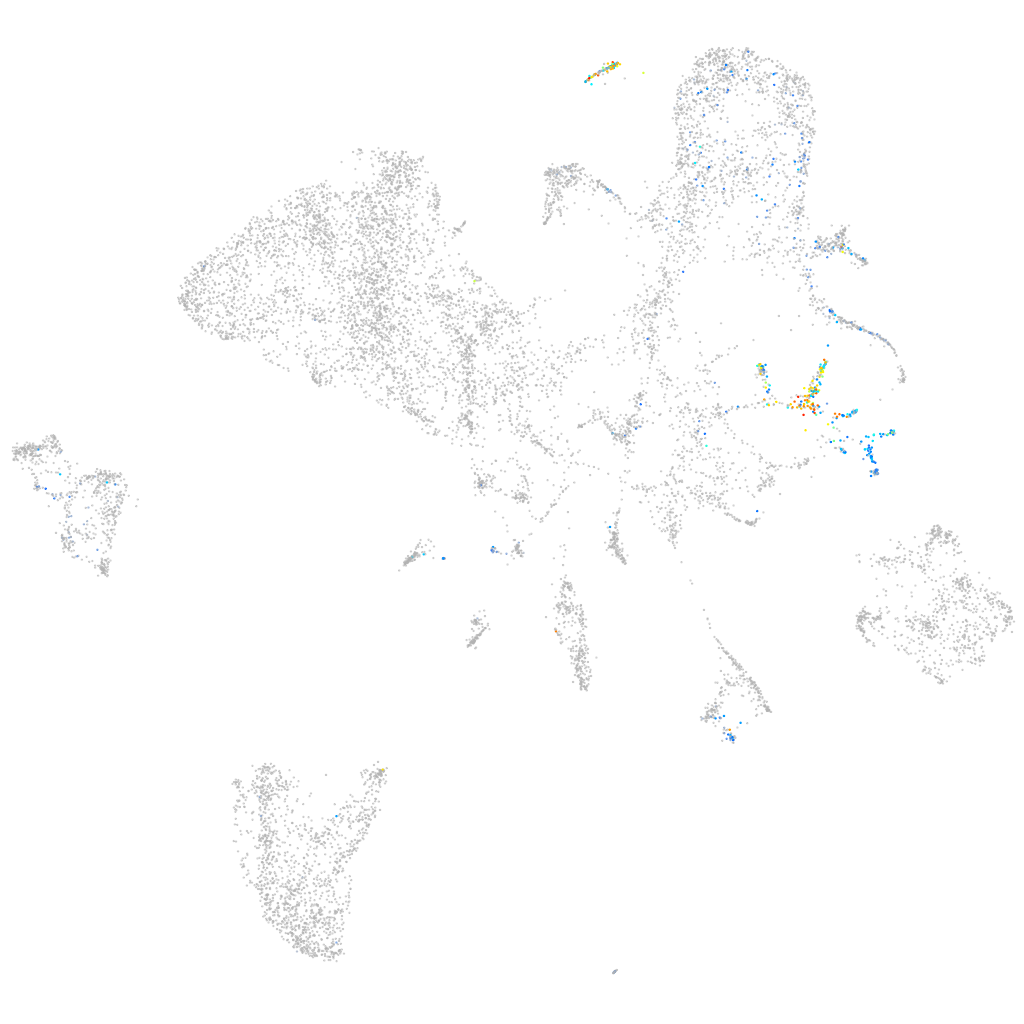
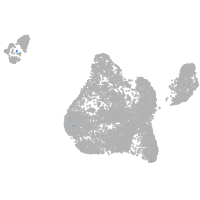
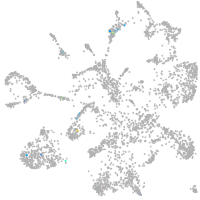
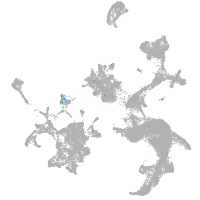
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.622 | fabp3 | -0.173 |
| neurod1 | 0.543 | ahcy | -0.171 |
| scgn | 0.518 | gamt | -0.162 |
| egr4 | 0.503 | gapdh | -0.159 |
| arxa | 0.497 | bhmt | -0.155 |
| pcsk1nl | 0.477 | gatm | -0.149 |
| fev | 0.471 | mat1a | -0.144 |
| zgc:101731 | 0.456 | aldh6a1 | -0.142 |
| syt1a | 0.450 | aqp12 | -0.138 |
| si:ch73-160i9.2 | 0.447 | apoc1 | -0.137 |
| insm1b | 0.445 | hdlbpa | -0.137 |
| rprmb | 0.442 | si:dkey-16p21.8 | -0.136 |
| pax6b | 0.430 | apoa1b | -0.131 |
| si:dkey-153k10.9 | 0.430 | agxtb | -0.131 |
| hepacam2 | 0.425 | apoa4b.1 | -0.130 |
| mir375-2 | 0.423 | agxta | -0.127 |
| pcsk2 | 0.420 | fbp1b | -0.126 |
| mir7a-1 | 0.417 | apoc2 | -0.124 |
| pcsk1 | 0.416 | cx32.3 | -0.124 |
| gpc1a | 0.414 | aldob | -0.123 |
| capsla | 0.411 | shmt1 | -0.123 |
| ghrl | 0.408 | aldh7a1 | -0.123 |
| nkx2.2a | 0.407 | hspe1 | -0.123 |
| tspan7b | 0.401 | rpl6 | -0.121 |
| rasd4 | 0.400 | afp4 | -0.121 |
| vat1 | 0.399 | ttc36 | -0.121 |
| kcnj11 | 0.390 | adka | -0.120 |
| chga | 0.383 | serpina1 | -0.120 |
| TCIM (1 of many) | 0.381 | gstt1a | -0.120 |
| tango2 | 0.380 | mgst1.2 | -0.120 |
| mapk15 | 0.379 | serpinf2b | -0.119 |
| insm1a | 0.374 | apoa2 | -0.119 |
| elovl1a | 0.370 | serpina1l | -0.119 |
| rims2a | 0.368 | cpn1 | -0.118 |
| isl1 | 0.366 | rps20 | -0.118 |