Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 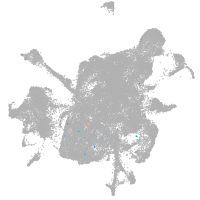 |
hematopoietic / vasculature  |
pronephros  |
neural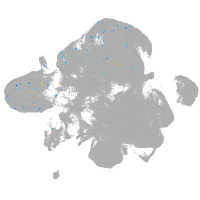 |
spinal cord / glia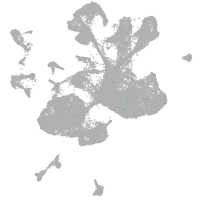 |
eye |
taste / olfactory |
otic / lateral line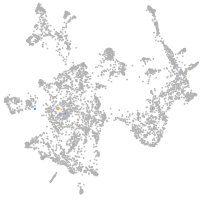 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmprss7 | 0.068 | stmn1b | -0.038 |
| LOC101882573 | 0.059 | elavl3 | -0.036 |
| pimr135 | 0.055 | rtn1a | -0.035 |
| LOC110439576 | 0.054 | ptmaa | -0.032 |
| slbp | 0.053 | gpm6ab | -0.032 |
| pcna | 0.052 | gng3 | -0.031 |
| chaf1a | 0.050 | ckbb | -0.031 |
| CDHR1 | 0.050 | gpm6aa | -0.031 |
| nasp | 0.050 | atp6v0cb | -0.030 |
| LOC101885413 | 0.049 | sncb | -0.030 |
| ccnd1 | 0.048 | tuba1c | -0.029 |
| XLOC-034181 | 0.048 | h2afx1 | -0.029 |
| dek | 0.047 | rtn1b | -0.029 |
| anp32b | 0.047 | tmsb | -0.028 |
| cdca7a | 0.046 | calm1a | -0.028 |
| ccne2 | 0.045 | rnasekb | -0.028 |
| mcm5 | 0.045 | elavl4 | -0.028 |
| mcm6 | 0.045 | vamp2 | -0.027 |
| hmgb2a | 0.044 | ywhah | -0.027 |
| rrm1 | 0.044 | atp6v1e1b | -0.027 |
| lig1 | 0.044 | atp6v1g1 | -0.027 |
| CR381579.4 | 0.044 | fez1 | -0.026 |
| ssrp1a | 0.044 | zc4h2 | -0.026 |
| banf1 | 0.043 | si:dkeyp-75h12.5 | -0.026 |
| zgc:110540 | 0.043 | jpt1b | -0.026 |
| cdc6 | 0.043 | stmn2a | -0.026 |
| cdca7b | 0.043 | ywhag2 | -0.026 |
| mcm3 | 0.043 | tmsb4x | -0.025 |
| hells | 0.043 | gapdhs | -0.025 |
| fen1 | 0.043 | ccni | -0.025 |
| CABZ01078244.2 | 0.042 | stx1b | -0.025 |
| fbxo5 | 0.041 | mllt11 | -0.025 |
| stmn1a | 0.041 | tubb5 | -0.025 |
| esco2 | 0.041 | CR383676.1 | -0.025 |
| tuba8l4 | 0.040 | stxbp1a | -0.025 |




