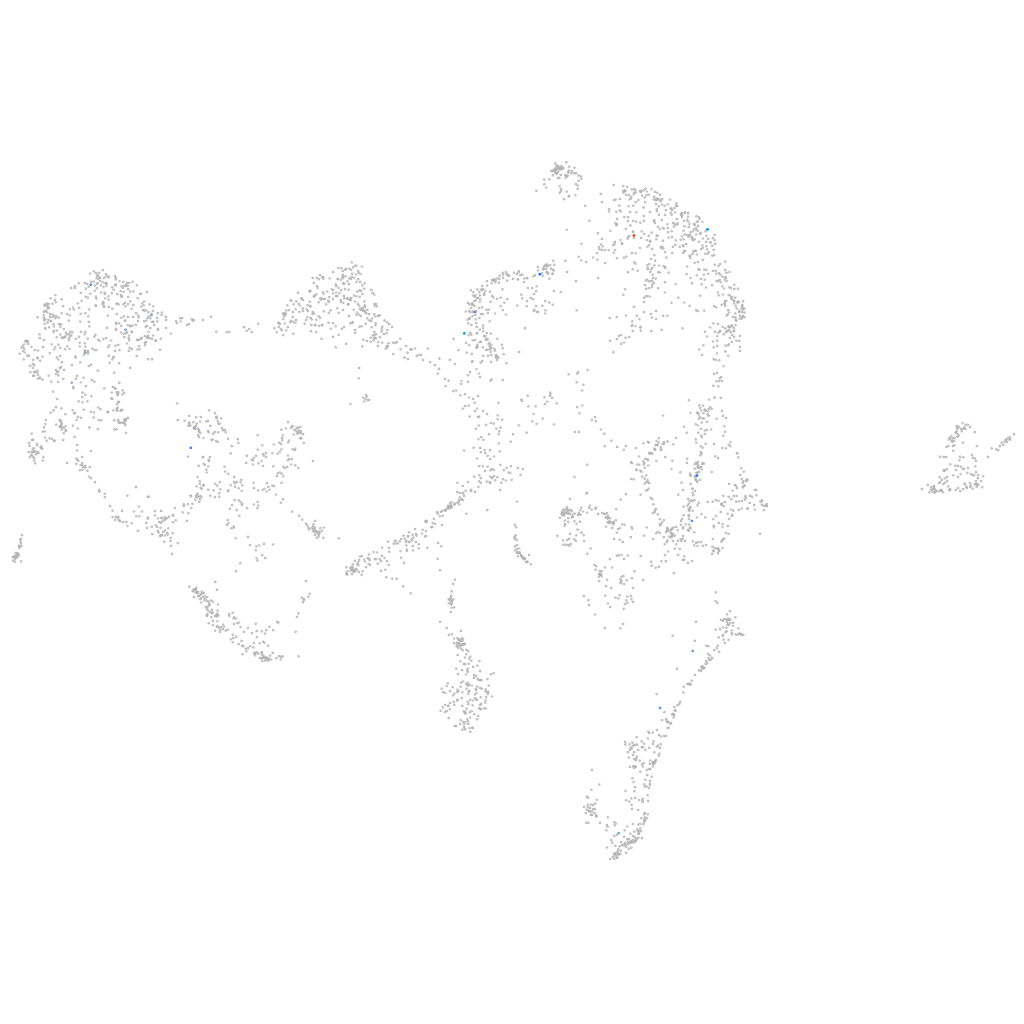
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-44a8.4 | 0.596 | rpl32 | -0.076 |
| si:ch73-167i17.6 | 0.591 | rpl17 | -0.065 |
| iqsec3a | 0.500 | slc25a5 | -0.065 |
| lrrc3b | 0.489 | rps12 | -0.062 |
| XLOC-017482 | 0.488 | rps9 | -0.060 |
| LOC101885528.1 | 0.372 | rpl29 | -0.056 |
| trim35-24 | 0.357 | rpl34 | -0.056 |
| pid1 | 0.345 | rps26 | -0.054 |
| CR381686.3 | 0.305 | naca | -0.046 |
| zbtb40 | 0.299 | nme2b.1 | -0.046 |
| bahcc1a | 0.282 | rps17 | -0.042 |
| BX005332.1 | 0.273 | cox6a1 | -0.041 |
| kcng1 | 0.273 | ndufa4l | -0.041 |
| mid2 | 0.272 | atp5f1e | -0.040 |
| snapc4 | 0.266 | cox4i1 | -0.040 |
| ly6m6 | 0.262 | cox6b2 | -0.037 |
| CR383662.4 | 0.251 | zgc:56493 | -0.036 |
| yrdc | 0.250 | cox7a2a | -0.036 |
| anapc2 | 0.249 | atp5f1d | -0.036 |
| specc1 | 0.241 | atp5pf | -0.035 |
| cyp2u1 | 0.240 | cox6c | -0.035 |
| CT990625.1 | 0.234 | cox7b | -0.034 |
| CR354383.1 | 0.232 | atp5mc1 | -0.033 |
| cuzd1.1 | 0.225 | cox7c | -0.032 |
| XLOC-033087 | 0.225 | atp5fa1 | -0.032 |
| si:ch211-230g15.5 | 0.220 | mgst3b | -0.032 |
| apom | 0.214 | rpl37 | -0.032 |
| nr1d1 | 0.212 | cox5ab | -0.032 |
| dusp10 | 0.211 | ola1 | -0.032 |
| ddx31 | 0.207 | rps23 | -0.031 |
| glsb | 0.202 | cfl1 | -0.031 |
| BX957362.1 | 0.200 | ndufa12 | -0.031 |
| bod1l1 | 0.196 | ube2e1 | -0.031 |
| marveld3 | 0.188 | atp5pd | -0.031 |
| slc7a2 | 0.186 | rpl5a | -0.031 |