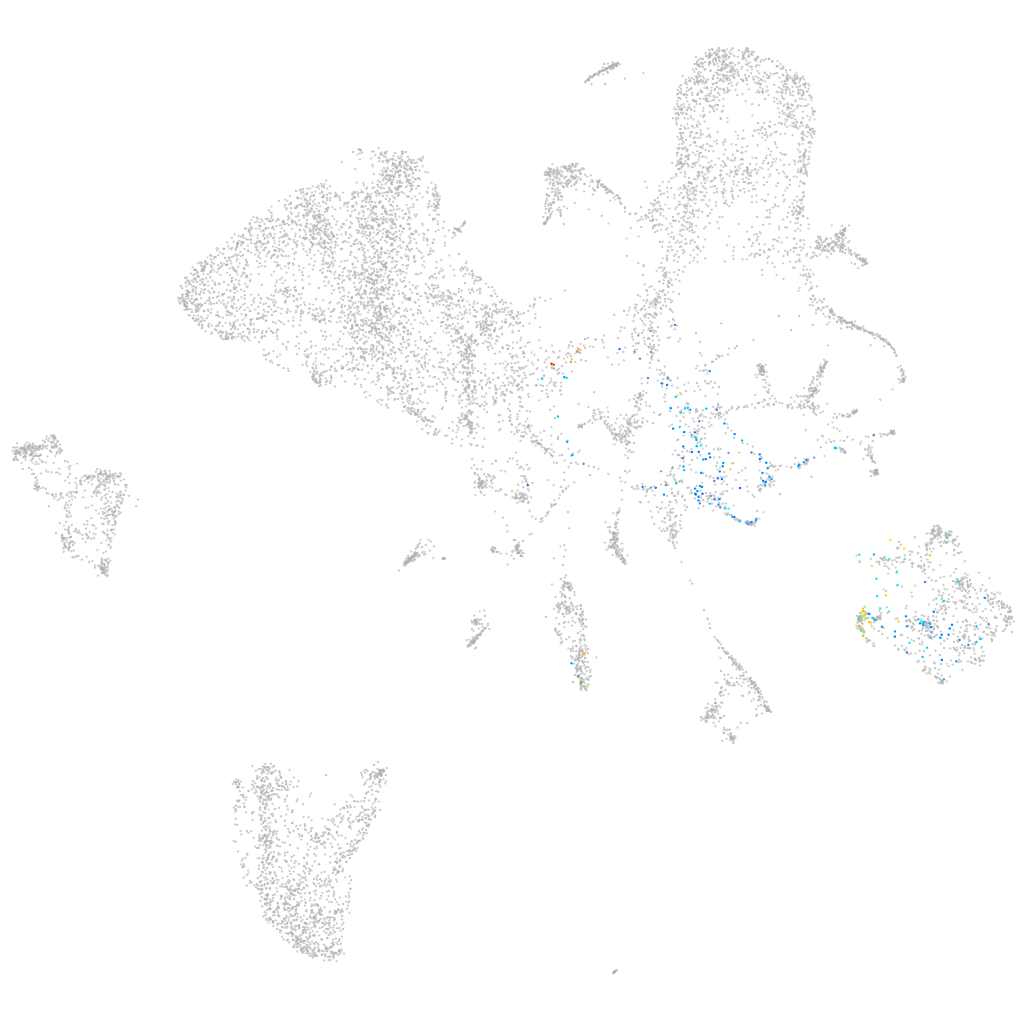
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mcama | 0.270 | nme2b.1 | -0.171 |
| BX469925.3 | 0.260 | sod1 | -0.159 |
| oc90 | 0.254 | ahcy | -0.156 |
| pltp | 0.242 | gapdh | -0.148 |
| cdh6 | 0.234 | eno3 | -0.147 |
| sept15 | 0.221 | rpl37 | -0.144 |
| CABZ01075068.1 | 0.221 | rps10 | -0.144 |
| fgfrl1b | 0.219 | atp5if1b | -0.134 |
| hspb1 | 0.218 | eef1da | -0.134 |
| marcksb | 0.211 | atp5mc1 | -0.134 |
| lsp1 | 0.205 | rps17 | -0.130 |
| id3 | 0.201 | zgc:114188 | -0.128 |
| crtap | 0.197 | gstp1 | -0.124 |
| zfp36l1a | 0.194 | glud1b | -0.122 |
| si:ch211-152c2.3 | 0.194 | atp5pf | -0.120 |
| qkia | 0.193 | suclg1 | -0.119 |
| tgif1 | 0.191 | cox6a1 | -0.119 |
| akap12b | 0.190 | atp5mc3b | -0.118 |
| si:ch73-281n10.2 | 0.187 | atp5f1b | -0.118 |
| tpm4a | 0.187 | gamt | -0.118 |
| hp1bp3 | 0.186 | mt-nd1 | -0.118 |
| hmgb3a | 0.184 | sod2 | -0.117 |
| syncrip | 0.183 | BX908782.3 | -0.116 |
| cx43.4 | 0.183 | nupr1b | -0.114 |
| NC-002333.4 | 0.182 | mt-atp6 | -0.114 |
| cd81a | 0.181 | zgc:158463 | -0.112 |
| marcksl1b | 0.181 | acadm | -0.111 |
| si:ch73-308l14.2 | 0.179 | pgk1 | -0.111 |
| apela | 0.179 | scp2a | -0.111 |
| zbtb16a | 0.178 | dap | -0.110 |
| smo | 0.178 | atp5fa1 | -0.110 |
| asph | 0.176 | cx32.3 | -0.110 |
| hmga1a | 0.175 | atp5pd | -0.110 |
| hmgn6 | 0.175 | prdx2 | -0.109 |
| nr6a1a | 0.175 | krtcap2 | -0.108 |