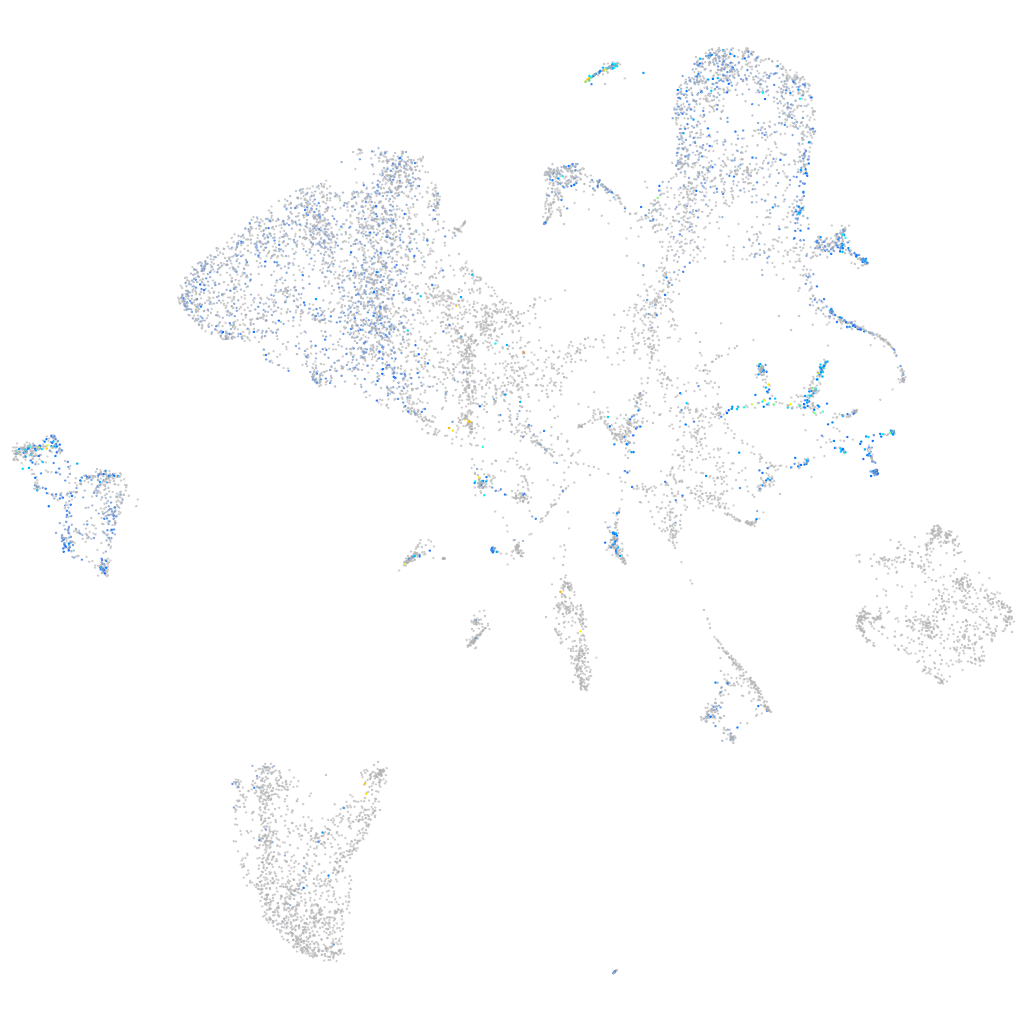
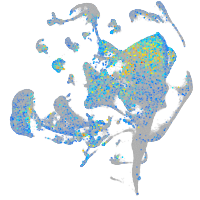
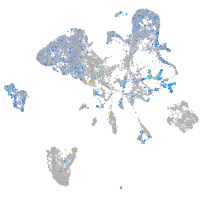
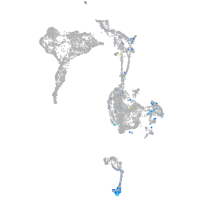
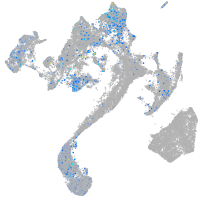
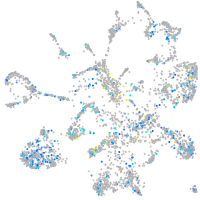
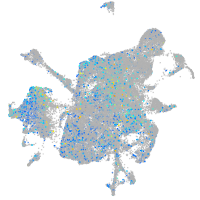
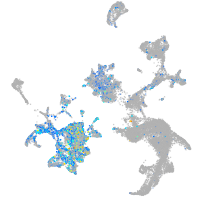
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.317 | npm1a | -0.131 |
| neurod1 | 0.275 | cpb1 | -0.126 |
| fev | 0.266 | ctrl | -0.125 |
| egr4 | 0.261 | cel.1 | -0.125 |
| pcsk1nl | 0.253 | fep15 | -0.124 |
| zgc:101731 | 0.250 | cel.2 | -0.124 |
| hepacam2 | 0.249 | cpa4 | -0.124 |
| mir375-2 | 0.249 | pdia2 | -0.123 |
| mir7a-1 | 0.248 | sycn.2 | -0.123 |
| c2cd4a | 0.236 | cpa5 | -0.122 |
| calm1b | 0.231 | ela2 | -0.122 |
| sat1a.2 | 0.230 | ela3l | -0.121 |
| cldnh | 0.228 | apoda.2 | -0.121 |
| dusp2 | 0.228 | zgc:112160 | -0.120 |
| adcyap1a | 0.228 | endou | -0.120 |
| pcsk1 | 0.226 | si:ch211-240l19.5 | -0.120 |
| dll4 | 0.217 | ela2l | -0.120 |
| insm1b | 0.215 | nop58 | -0.120 |
| scg2a | 0.215 | ctrb1 | -0.119 |
| ier2b | 0.210 | amy2a | -0.118 |
| syt1a | 0.207 | zgc:136461 | -0.118 |
| elovl1a | 0.207 | prss1 | -0.117 |
| scgn | 0.207 | aqp12 | -0.117 |
| acbd7 | 0.205 | prss59.1 | -0.116 |
| zgc:165481 | 0.203 | prss59.2 | -0.116 |
| trpa1b | 0.203 | erp27 | -0.116 |
| nkx2.2a | 0.203 | c6ast4 | -0.116 |
| pax6b | 0.202 | CELA1 (1 of many) | -0.116 |
| nr4a1 | 0.200 | zgc:92137 | -0.116 |
| calm1a | 0.200 | cpa1 | -0.115 |
| capsla | 0.199 | si:ch211-240l19.6 | -0.113 |
| tmem59 | 0.198 | fbl | -0.111 |
| gapdhs | 0.197 | tmed6 | -0.110 |
| gpc1a | 0.195 | si:ch73-44m9.5 | -0.110 |
| rnasekb | 0.191 | hspb1 | -0.107 |