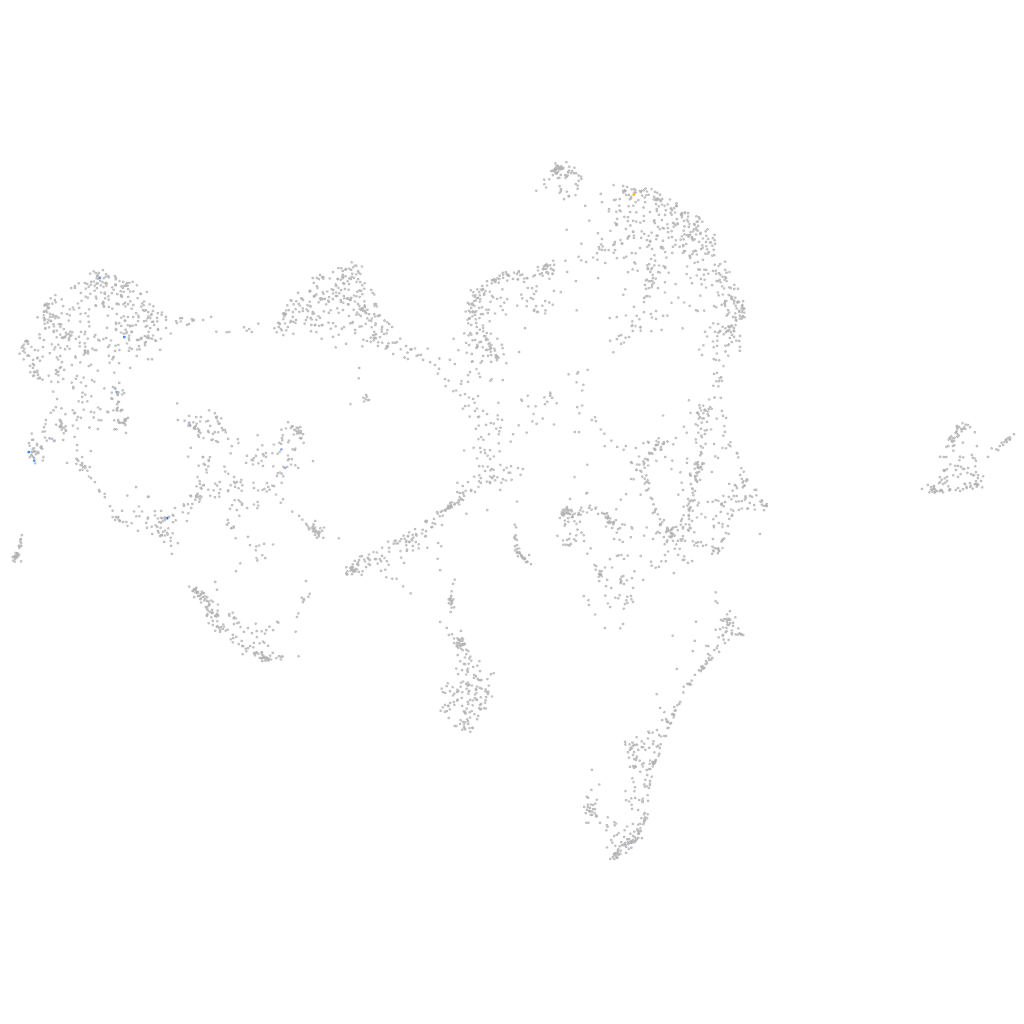
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc6a19a.1 | 0.878 | rpl23a | -0.056 |
| si:ch211-125m10.6 | 0.851 | mt-atp6 | -0.045 |
| cfh | 0.652 | rps17 | -0.045 |
| lenep | 0.569 | rpl3 | -0.039 |
| slc2a2 | 0.529 | cox8a | -0.037 |
| dnajc16 | 0.521 | mt-nd1 | -0.037 |
| cobl | 0.503 | COX3 | -0.037 |
| LOC110440168 | 0.497 | rplp2l | -0.036 |
| hoxa13b | 0.457 | NC-002333.17 | -0.035 |
| adat1 | 0.401 | ndufa4l | -0.035 |
| LOC103911430 | 0.397 | rps18 | -0.034 |
| LO018166.1 | 0.388 | NC-002333.4 | -0.033 |
| dennd4b | 0.316 | hmgn6 | -0.032 |
| smyd1a | 0.302 | btg1 | -0.031 |
| ASTE1 | 0.298 | COX5B | -0.031 |
| b3gnt2a | 0.286 | atp5fa1 | -0.031 |
| scarb1 | 0.275 | cox6a1 | -0.031 |
| pklr | 0.260 | mt-nd2 | -0.030 |
| CABZ01061524.1 | 0.243 | atp5f1d | -0.030 |
| mier2 | 0.236 | tmed2 | -0.030 |
| rab12 | 0.234 | rpl5b | -0.030 |
| myom1a | 0.232 | ppib | -0.030 |
| cpb2 | 0.222 | atp5f1b | -0.029 |
| LOC103910947 | 0.213 | cfl1 | -0.029 |
| agxtb | 0.211 | tmem258 | -0.029 |
| prmt3 | 0.207 | s100u | -0.029 |
| rsf1a | 0.205 | cox7c | -0.029 |
| serpina1l | 0.198 | atp5pd | -0.029 |
| trib2 | 0.198 | atp5f1e | -0.029 |
| plekhm1 | 0.197 | ldhba | -0.028 |
| kank4 | 0.185 | cox5aa | -0.028 |
| apom | 0.181 | ywhaqb | -0.028 |
| serac1 | 0.180 | atp5meb | -0.028 |
| ces2 | 0.174 | zgc:193541 | -0.028 |
| pxylp1 | 0.173 | atp5pf | -0.028 |