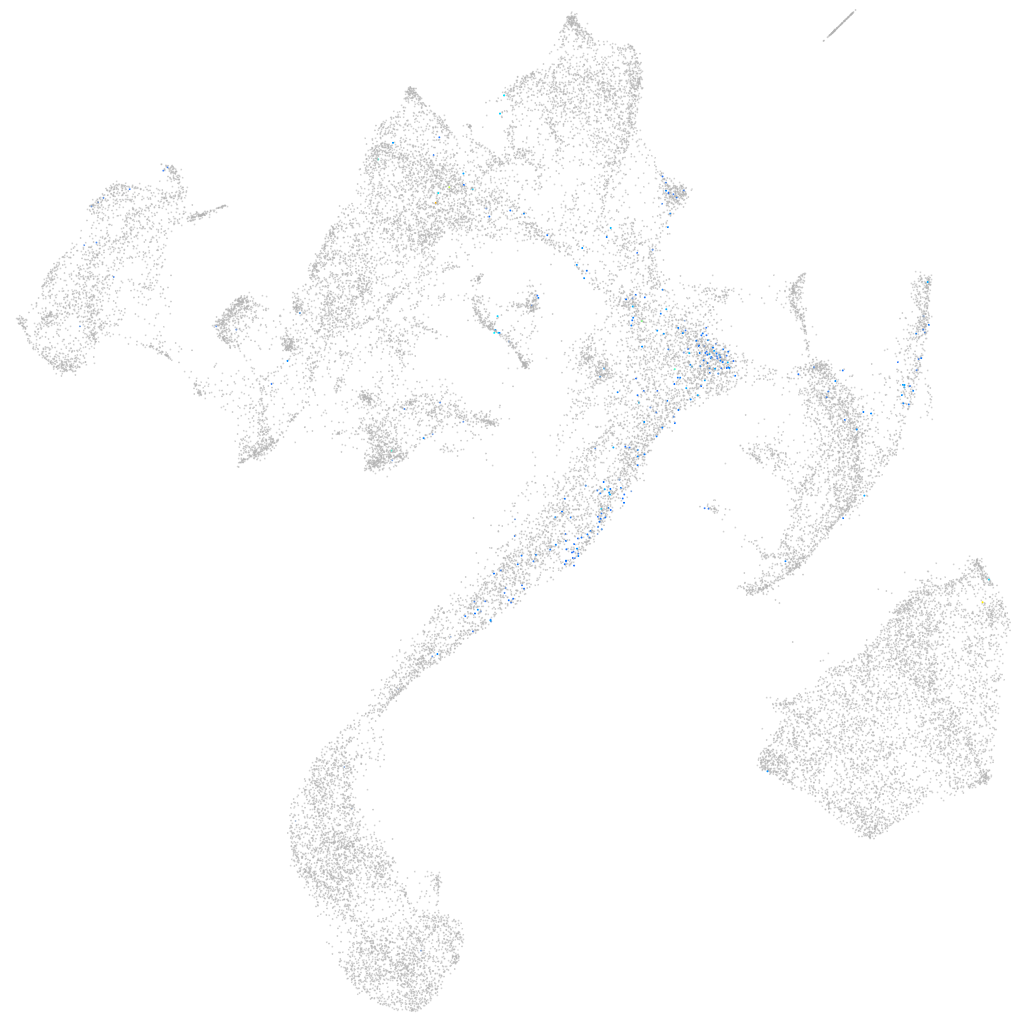
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL929396.1 | 0.098 | si:dkey-16p21.8 | -0.048 |
| CU695215.2 | 0.092 | pvalb1 | -0.045 |
| XLOC-042229 | 0.092 | pvalb2 | -0.044 |
| fstl1a | 0.092 | bhmt | -0.042 |
| XLOC-037389 | 0.089 | si:ch211-152c2.3 | -0.042 |
| efemp2a | 0.089 | tnnt3b | -0.041 |
| bmpr1ba | 0.088 | cdx4 | -0.040 |
| plpp1a | 0.084 | ckmb | -0.039 |
| fn1b | 0.084 | ckma | -0.039 |
| mafbb | 0.084 | si:ch73-367p23.2 | -0.039 |
| fshb | 0.083 | tnni2a.4 | -0.039 |
| CABZ01025311.1 | 0.082 | ndrg2 | -0.038 |
| lman1 | 0.082 | prx | -0.038 |
| zgc:92429 | 0.080 | actn3a | -0.038 |
| kazald2 | 0.076 | tnnt3a | -0.038 |
| cpn1 | 0.076 | rps18 | -0.038 |
| CT573231.1 | 0.075 | eno3 | -0.038 |
| btf3 | 0.073 | glud1b | -0.037 |
| LOC103910947 | 0.072 | myom1a | -0.037 |
| ripply1 | 0.072 | cox7c | -0.037 |
| si:dkey-68o6.8 | 0.072 | pgam2 | -0.037 |
| zbtb18 | 0.071 | mylz3 | -0.036 |
| thrap3a | 0.071 | gapdh | -0.036 |
| btf3l4 | 0.071 | si:ch211-266g18.10 | -0.036 |
| sirt7 | 0.071 | unm-hu7910 | -0.036 |
| angptl7 | 0.070 | ldb3b | -0.036 |
| si:dkeyp-68b7.12 | 0.070 | XLOC-006515 | -0.035 |
| BX119915.3 | 0.070 | casq1b | -0.035 |
| fam49a | 0.070 | cavin4a | -0.035 |
| emg1 | 0.069 | smyd1a | -0.035 |
| fam107b | 0.069 | XLOC-001975 | -0.035 |
| aktip | 0.069 | XLOC-025819 | -0.035 |
| XLOC-029508 | 0.069 | tmod4 | -0.035 |
| optc | 0.068 | si:ch211-255p10.3 | -0.035 |
| vaspb | 0.068 | vox | -0.035 |