Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 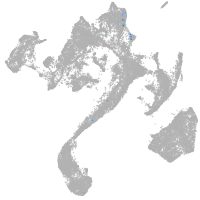 |
mural cells / non-skeletal muscle 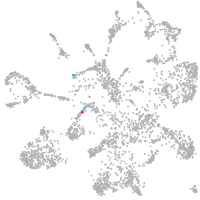 |
mesenchyme 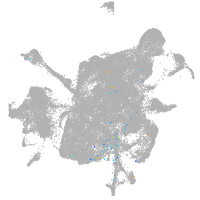 |
hematopoietic / vasculature 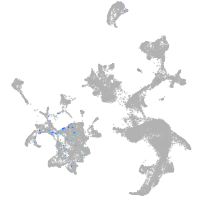 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nkx3.2 | 0.058 | gpm6aa | -0.015 |
| foxf2a | 0.057 | tuba1c | -0.015 |
| ramp2 | 0.057 | fabp3 | -0.014 |
| foxf2b | 0.055 | cadm3 | -0.013 |
| egfl7 | 0.052 | ckbb | -0.013 |
| si:ch73-86n18.1 | 0.052 | si:ch211-137a8.4 | -0.013 |
| rasip1 | 0.051 | fabp7a | -0.012 |
| cpn1 | 0.050 | gpm6ab | -0.012 |
| etv2 | 0.050 | rtn1a | -0.012 |
| clec14a | 0.049 | tuba1a | -0.012 |
| stab2 | 0.049 | CU467822.1 | -0.011 |
| apof | 0.048 | cyt1 | -0.011 |
| itga11a | 0.048 | elavl3 | -0.011 |
| tnfaip2b | 0.048 | krt4 | -0.011 |
| cfi | 0.047 | stmn1b | -0.011 |
| gpr182 | 0.047 | CU634008.1 | -0.011 |
| si:dkeyp-106c3.1 | 0.047 | cspg5a | -0.010 |
| colec12 | 0.046 | epb41a | -0.010 |
| foxf1 | 0.046 | epcam | -0.010 |
| tgfbi | 0.046 | krtt1c19e | -0.010 |
| myct1a | 0.045 | myt1b | -0.010 |
| reck | 0.045 | nova2 | -0.010 |
| mrc1a | 0.045 | tmsb | -0.010 |
| flt4 | 0.044 | anxa1a | -0.009 |
| FO704741.1 | 0.044 | cfl1l | -0.009 |
| scarf1 | 0.044 | fez1 | -0.009 |
| si:dkey-52l18.4 | 0.044 | gng3 | -0.009 |
| si:dkeyp-97a10.2 | 0.044 | icn | -0.009 |
| tpm4a | 0.044 | mab21l1 | -0.009 |
| erg | 0.043 | myt1a | -0.009 |
| twist2 | 0.043 | pou3f3b | -0.009 |
| cdh5 | 0.042 | rnasekb | -0.009 |
| foxd2 | 0.042 | rtn1b | -0.009 |
| she | 0.042 | si:dkey-248g15.3 | -0.009 |
| zgc:162612 | 0.042 | si:dkey-276j7.1 | -0.009 |




