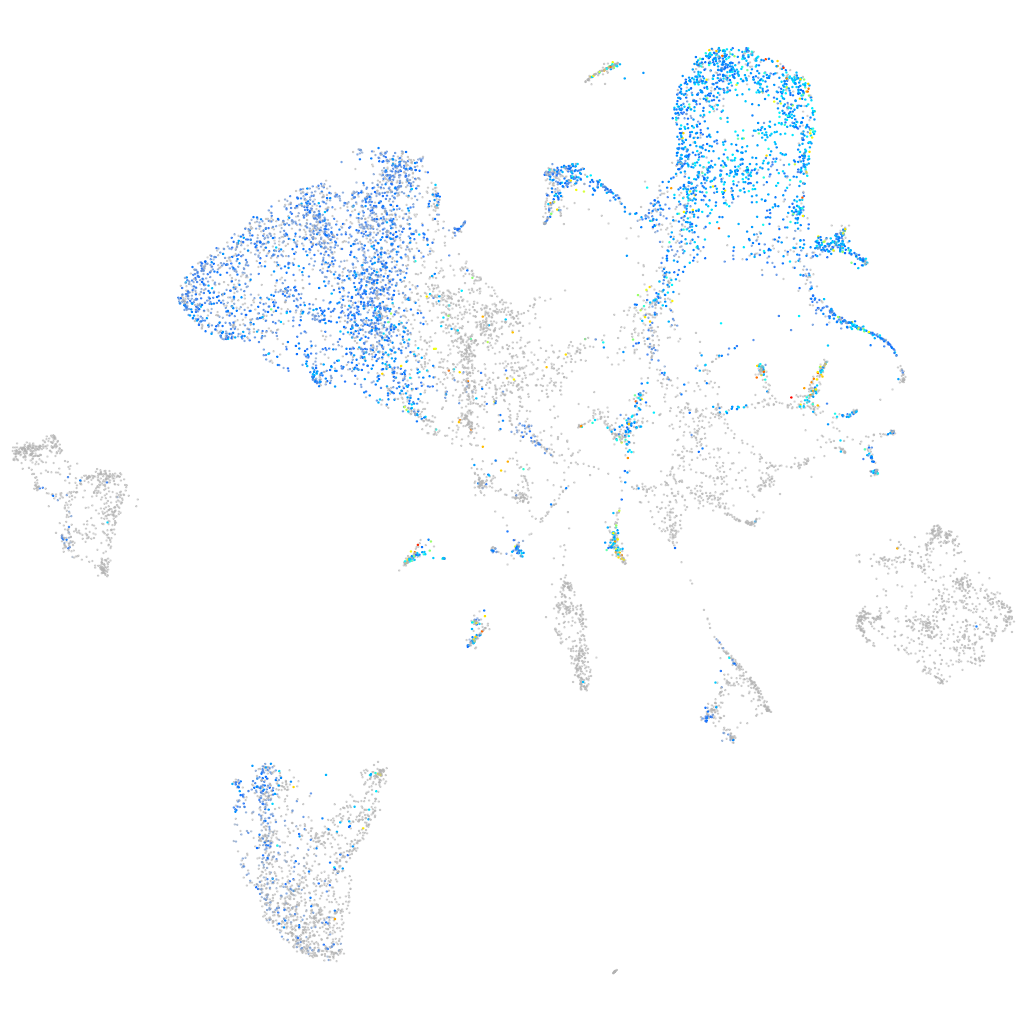
Other cell groups


all cells |
blastomeres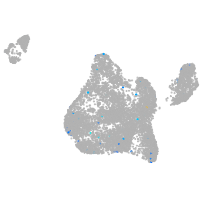 |
PGCs |
endoderm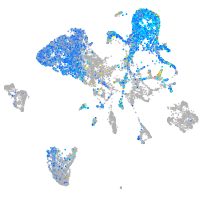 |
axial mesoderm |
muscle 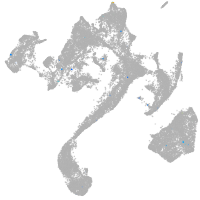 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-73b11.8 | 0.497 | cx43.4 | -0.218 |
| lgals2b | 0.497 | marcksb | -0.212 |
| vil1 | 0.489 | hspb1 | -0.207 |
| atp1a1a.4 | 0.479 | marcksl1b | -0.204 |
| si:dkey-36i7.3 | 0.466 | akap12b | -0.203 |
| zgc:172079 | 0.464 | qkia | -0.199 |
| zgc:158846 | 0.464 | glulb | -0.185 |
| si:ch211-133l5.7 | 0.464 | cdh6 | -0.183 |
| AL831745.1 | 0.462 | slc38a5b | -0.182 |
| cdh17 | 0.460 | si:ch211-195b11.3 | -0.182 |
| ace2 | 0.458 | msna | -0.179 |
| cldn15la | 0.457 | si:ch211-152c2.3 | -0.178 |
| ada | 0.456 | stm | -0.173 |
| zanl | 0.455 | cd81a | -0.170 |
| dhrs1 | 0.455 | tpm4a | -0.167 |
| tm4sf4 | 0.453 | ctrl | -0.167 |
| s100a10a | 0.452 | hmgb1b | -0.164 |
| abcg2a | 0.448 | id1 | -0.161 |
| slc6a19a.2 | 0.448 | rtn1a | -0.159 |
| zgc:153968 | 0.446 | tgif1 | -0.159 |
| anxa2b | 0.446 | ela2l | -0.157 |
| ugt1a7 | 0.443 | smo | -0.156 |
| si:ch211-107o10.3 | 0.439 | sycn.2 | -0.156 |
| muc13b | 0.439 | ackr3b | -0.156 |
| plac8.1 | 0.438 | c6ast4 | -0.155 |
| calml4a | 0.435 | cpa4 | -0.155 |
| aqp8a.2 | 0.432 | zgc:112160 | -0.155 |
| itpk1a | 0.432 | fabp3 | -0.155 |
| wu:fb59d01 | 0.428 | zgc:136461 | -0.154 |
| pdzk1 | 0.428 | ela2 | -0.154 |
| slc15a1b | 0.426 | prss59.1 | -0.153 |
| zgc:77748 | 0.425 | cpb1 | -0.153 |
| abcc2 | 0.425 | ctrb1 | -0.153 |
| mansc1 | 0.425 | lamb1a | -0.153 |
| serpinb1l3 | 0.425 | ela3l | -0.153 |