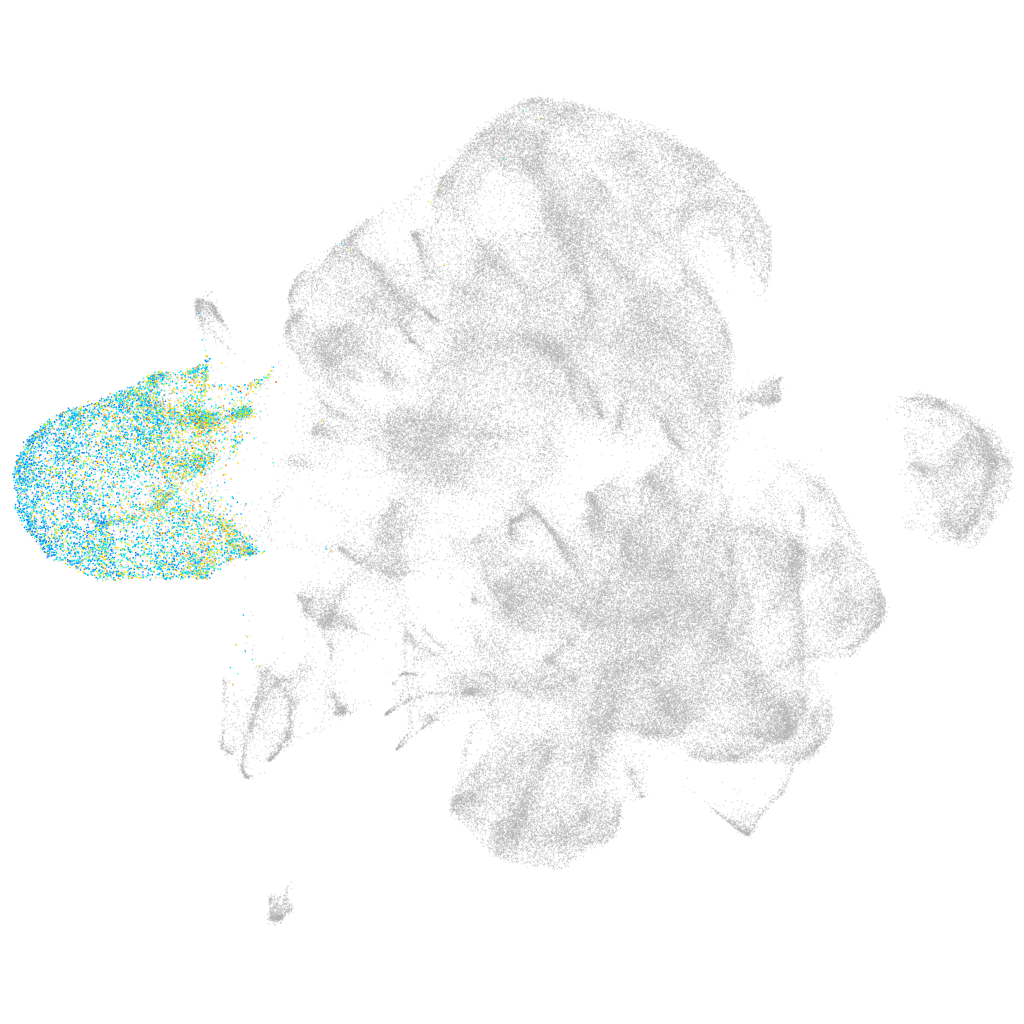
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152c2.3 | 0.590 | rps10 | -0.500 |
| hspb1 | 0.572 | rpl37 | -0.497 |
| apoeb | 0.532 | zgc:114188 | -0.471 |
| stm | 0.527 | h3f3a | -0.448 |
| NC-002333.4 | 0.518 | hnrnpa0l | -0.412 |
| apoc1 | 0.484 | ptmaa | -0.398 |
| zmp:0000000624 | 0.462 | rps17 | -0.360 |
| crabp2b | 0.458 | fabp3 | -0.347 |
| nr6a1a | 0.452 | marcksl1a | -0.342 |
| fbl | 0.447 | CR383676.1 | -0.316 |
| pou5f3 | 0.437 | ppiab | -0.300 |
| si:dkey-66i24.9 | 0.435 | tuba1c | -0.299 |
| polr3gla | 0.434 | si:ch1073-429i10.3.1 | -0.299 |
| dkc1 | 0.426 | tmsb4x | -0.286 |
| s100a1 | 0.420 | cct2 | -0.275 |
| gnl3 | 0.415 | zgc:158463 | -0.271 |
| gar1 | 0.414 | nme2b.1 | -0.260 |
| npm1a | 0.414 | h3f3c | -0.238 |
| nop58 | 0.413 | mt-nd1 | -0.238 |
| bms1 | 0.413 | elavl3 | -0.237 |
| shisa2a | 0.411 | zgc:56493 | -0.236 |
| nop2 | 0.411 | rtn1a | -0.230 |
| ncl | 0.408 | gpm6aa | -0.229 |
| apela | 0.407 | nova2 | -0.217 |
| ddx18 | 0.403 | eef1b2 | -0.215 |
| anp32e | 0.402 | wu:fb55g09 | -0.211 |
| ISCU | 0.395 | stmn1b | -0.210 |
| alcamb | 0.394 | zgc:153409 | -0.208 |
| rbmx2 | 0.394 | celf2 | -0.208 |
| pkdccb | 0.393 | ppdpfb | -0.205 |
| aldob | 0.386 | fam168a | -0.202 |
| hnrnpa1b | 0.386 | calm1a | -0.201 |
| rrp15 | 0.385 | prdx2 | -0.200 |
| ebna1bp2 | 0.385 | tmsb | -0.198 |
| rsl1d1 | 0.384 | NC-002333.17 | -0.195 |