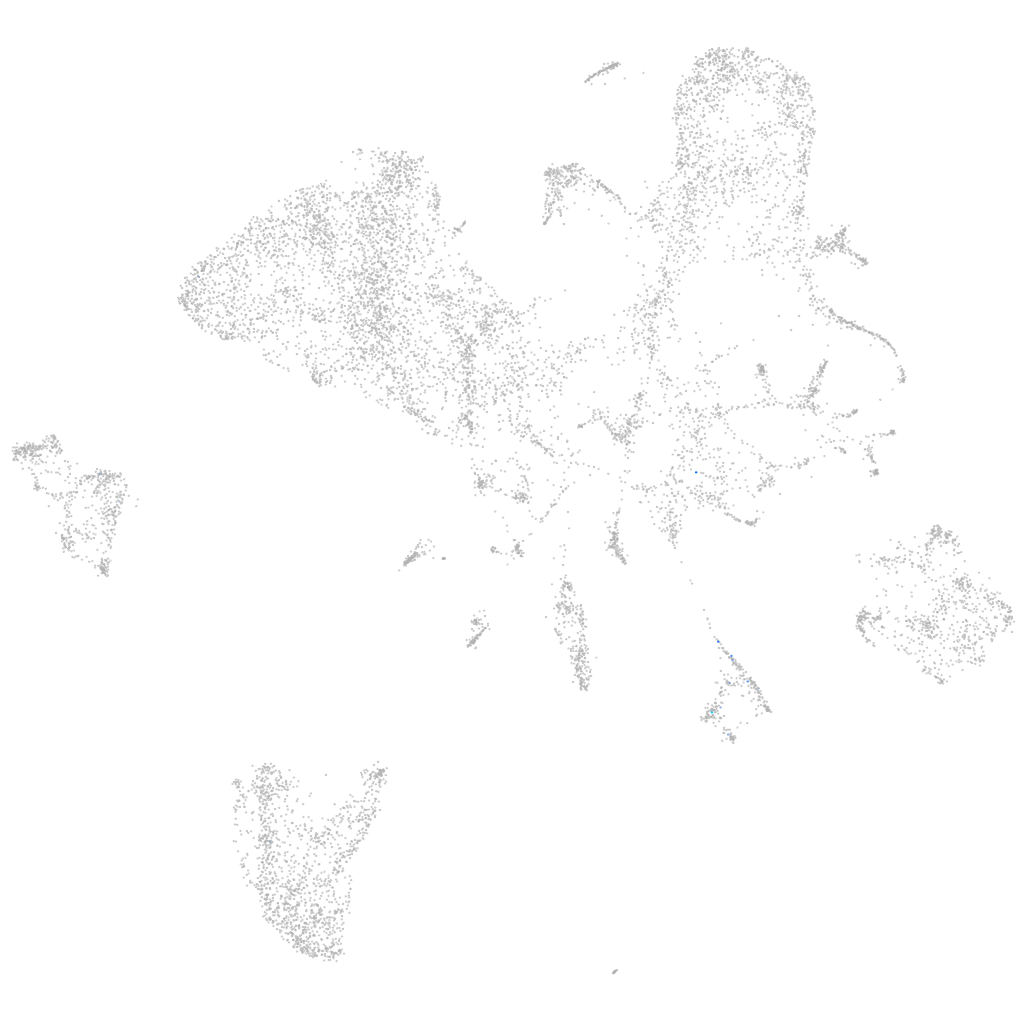
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX547934.2 | 0.509 | lgals2b | -0.026 |
| zmp:0000001132 | 0.331 | fbp1b | -0.025 |
| CR847899.2 | 0.258 | apoa4b.1 | -0.025 |
| sema5ba | 0.249 | gpx4a | -0.025 |
| si:dkeyp-104f11.8 | 0.226 | apoa1b | -0.025 |
| sox21b | 0.218 | apoc2 | -0.025 |
| cdc42ep1b | 0.187 | afp4 | -0.024 |
| trim35-1 | 0.181 | gapdh | -0.024 |
| vwa7 | 0.180 | pklr | -0.024 |
| c1qtnf2 | 0.175 | gstt1a | -0.023 |
| cxcl11.1 | 0.168 | gcshb | -0.023 |
| etv7 | 0.164 | ugt1a7 | -0.023 |
| LOC110440131 | 0.159 | bhmt | -0.023 |
| vwa10.2 | 0.149 | gatm | -0.022 |
| CABZ01075572.1 | 0.147 | gamt | -0.022 |
| sim1b | 0.145 | mat1a | -0.022 |
| loxa | 0.143 | cldn15lb | -0.022 |
| LOC101886229 | 0.141 | sb:cb1058 | -0.022 |
| CU459145.1 | 0.136 | apoc1 | -0.021 |
| frem2b | 0.135 | lipf | -0.021 |
| LOC101882402 | 0.135 | suclg2 | -0.021 |
| CU655961.1 | 0.133 | dhrs9 | -0.021 |
| rsph10b | 0.131 | scp2a | -0.021 |
| cabp2b | 0.129 | sult2st2 | -0.021 |
| si:ch211-157p22.10 | 0.117 | si:dkeyp-73b11.8 | -0.021 |
| osbpl1a | 0.113 | acmsd | -0.021 |
| spock3 | 0.113 | g6pca.2 | -0.021 |
| sftpba | 0.111 | gnmt | -0.021 |
| ankrd34bb | 0.111 | clic5b | -0.021 |
| CABZ01040055.1 | 0.110 | si:dkey-16p21.8 | -0.021 |
| vwa1 | 0.110 | aldh8a1 | -0.020 |
| actn1 | 0.110 | rdh1 | -0.020 |
| parp12a | 0.109 | mt-nd3 | -0.020 |
| pkd2l1 | 0.109 | apoa2 | -0.020 |
| ugt2b3 | 0.108 | tdo2a | -0.020 |