Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 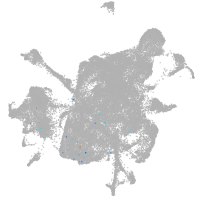 |
hematopoietic / vasculature 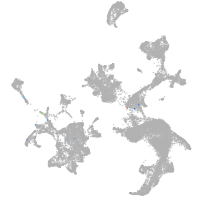 |
pronephros 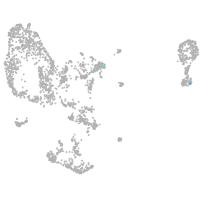 |
neural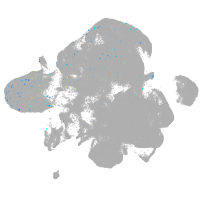 |
spinal cord / glia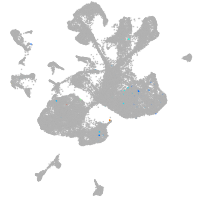 |
eye |
taste / olfactory |
otic / lateral line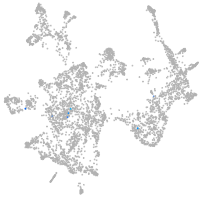 |
epidermis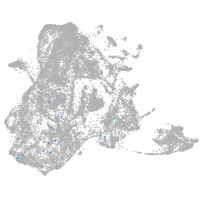 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nusap1 | 0.062 | ptmaa | -0.044 |
| CR318620.1 | 0.059 | rtn1a | -0.039 |
| mki67 | 0.058 | tuba1c | -0.038 |
| lck | 0.058 | stmn1b | -0.037 |
| ube2c | 0.056 | CR383676.1 | -0.035 |
| mad2l1 | 0.054 | gng3 | -0.035 |
| tpx2 | 0.053 | elavl3 | -0.034 |
| plk1 | 0.052 | gpm6ab | -0.034 |
| hmgb2a | 0.052 | gpm6aa | -0.034 |
| ccnb1 | 0.052 | sncb | -0.033 |
| dkc1 | 0.051 | tmsb | -0.033 |
| BX324137.1 | 0.051 | ckbb | -0.032 |
| dlgap5 | 0.050 | rnasekb | -0.032 |
| nop2 | 0.050 | calm1a | -0.032 |
| si:ch73-281n10.2 | 0.050 | atp6v0cb | -0.031 |
| cenpf | 0.049 | rtn1b | -0.031 |
| cdca8 | 0.049 | atp6v1e1b | -0.030 |
| nop58 | 0.049 | vamp2 | -0.030 |
| banf1 | 0.049 | ppdpfb | -0.030 |
| kmt5ab | 0.048 | tmsb4x | -0.030 |
| BX321882.1 | 0.048 | stmn2a | -0.029 |
| kif11 | 0.048 | rpl37 | -0.029 |
| si:ch211-69g19.2 | 0.047 | ywhag2 | -0.029 |
| hspb1 | 0.047 | ywhah | -0.029 |
| npm1a | 0.047 | fez1 | -0.029 |
| akap12b | 0.047 | atp6v1g1 | -0.028 |
| birc5a | 0.046 | gapdhs | -0.028 |
| aurka | 0.046 | elavl4 | -0.028 |
| aspm | 0.046 | ppiab | -0.028 |
| nop56 | 0.045 | celf2 | -0.028 |
| ccna2 | 0.045 | stxbp1a | -0.028 |
| pprc1 | 0.045 | stx1b | -0.028 |
| zgc:110425 | 0.045 | vdac3 | -0.027 |
| si:ch211-152c2.3 | 0.044 | gap43 | -0.027 |
| kif22 | 0.044 | COX3 | -0.027 |




