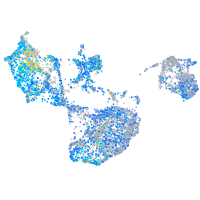
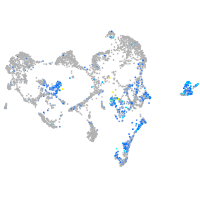
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cx43.4 | 0.478 | actc1b | -0.473 |
| si:ch211-222l21.1 | 0.457 | ckmb | -0.433 |
| ptmab | 0.456 | atp2a1 | -0.432 |
| si:ch73-1a9.3 | 0.455 | ak1 | -0.430 |
| si:ch73-281n10.2 | 0.445 | aldoab | -0.429 |
| nucks1a | 0.432 | ckma | -0.428 |
| cirbpa | 0.432 | tmem38a | -0.423 |
| hnrnpaba | 0.430 | pabpc4 | -0.416 |
| apoc1 | 0.426 | ttn.2 | -0.410 |
| hmga1a | 0.426 | tnnc2 | -0.406 |
| hmgb2a | 0.424 | acta1b | -0.404 |
| khdrbs1a | 0.423 | neb | -0.393 |
| pabpc1a | 0.420 | mylpfa | -0.391 |
| hmgn7 | 0.417 | gamt | -0.391 |
| hsp90ab1 | 0.413 | gapdh | -0.390 |
| apoeb | 0.413 | tpma | -0.389 |
| hmgn6 | 0.412 | CABZ01078594.1 | -0.388 |
| syncrip | 0.412 | srl | -0.388 |
| hmgb2b | 0.411 | mybphb | -0.385 |
| acin1a | 0.408 | ldb3b | -0.385 |
| h2afvb | 0.407 | eno1a | -0.380 |
| h3f3d | 0.407 | ldb3a | -0.379 |
| seta | 0.406 | myl1 | -0.379 |
| hes6 | 0.403 | si:ch73-367p23.2 | -0.373 |
| anp32a | 0.402 | actn3a | -0.372 |
| cirbpb | 0.398 | cav3 | -0.370 |
| anp32b | 0.397 | desma | -0.370 |
| si:ch211-288g17.3 | 0.395 | ttn.1 | -0.368 |
| pnrc2 | 0.394 | nme2b.2 | -0.367 |
| ubc | 0.393 | mylz3 | -0.366 |
| hnrnpabb | 0.389 | tnnt3a | -0.366 |
| nid2a | 0.386 | ank1a | -0.363 |
| marcksb | 0.385 | myom1a | -0.362 |
| hdac1 | 0.381 | actn3b | -0.360 |
| ctnnb1 | 0.378 | hhatla | -0.358 |