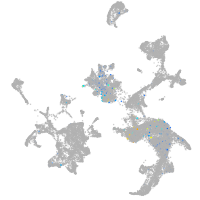
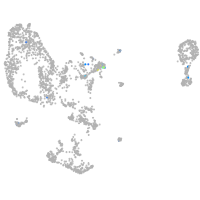
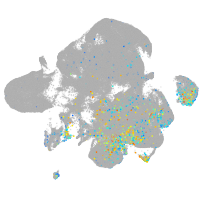
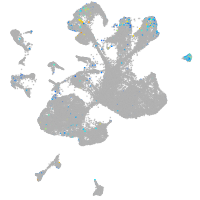
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.374 | gapdh | -0.115 |
| neurod1 | 0.371 | aldob | -0.115 |
| mir7a-1 | 0.363 | ahcy | -0.109 |
| pax6b | 0.326 | gstr | -0.104 |
| syt1a | 0.323 | gstp1 | -0.098 |
| id4 | 0.309 | apoa4b.1 | -0.098 |
| si:dkey-153k10.9 | 0.308 | gstt1a | -0.097 |
| insm1a | 0.307 | apoc2 | -0.096 |
| pcsk1nl | 0.295 | fbp1b | -0.096 |
| pygma | 0.286 | gamt | -0.094 |
| mir375-2 | 0.286 | apoa1b | -0.091 |
| ndufa4l2a | 0.283 | afp4 | -0.087 |
| scgn | 0.283 | apobb.1 | -0.087 |
| scg2a | 0.283 | ckba | -0.087 |
| kcnj11 | 0.273 | gcshb | -0.087 |
| rprmb | 0.272 | apoc1 | -0.085 |
| nkx2.2a | 0.270 | suclg2 | -0.085 |
| isl1 | 0.267 | scp2a | -0.084 |
| vamp2 | 0.266 | ugt1a7 | -0.084 |
| lysmd2 | 0.260 | rmdn1 | -0.084 |
| kcnk9 | 0.257 | mgst1.2 | -0.083 |
| vwa5b2 | 0.251 | si:dkey-16p21.8 | -0.083 |
| vat1 | 0.250 | aldh7a1 | -0.083 |
| gpc1a | 0.250 | aldh6a1 | -0.083 |
| zgc:101731 | 0.250 | aldh1l1 | -0.082 |
| egr4 | 0.249 | sult2st2 | -0.080 |
| fev | 0.247 | fabp2 | -0.080 |
| myt1b | 0.245 | ppa1b | -0.080 |
| foxo1a | 0.245 | gatm | -0.079 |
| cacna1c | 0.245 | shmt1 | -0.079 |
| spon1b | 0.242 | g6pca.2 | -0.078 |
| tmem63c | 0.241 | tdo2a | -0.078 |
| atp6v0cb | 0.238 | apoa2 | -0.078 |
| insm1b | 0.237 | gpx4a | -0.077 |
| dll4 | 0.237 | CABZ01032488.1 | -0.077 |